MARCKS and PI(4,5)P2 reciprocally regulate actin-based dendritic spine morphology
- PMID: 38088877
- PMCID: PMC10881156
- DOI: 10.1091/mbc.E23-09-0370
MARCKS and PI(4,5)P2 reciprocally regulate actin-based dendritic spine morphology
Abstract
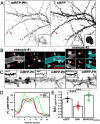
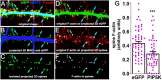
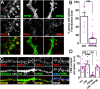
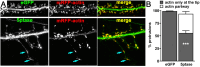
Myristoylated, alanine-rich C-kinase substrate (MARCKS) is an F-actin and phospholipid binding protein implicated in numerous cellular activities, including the regulation of morphology in neuronal dendrites and dendritic spines. MARCKS contains a lysine-rich effector domain that mediates its binding to plasma membrane phosphatidylinositol-4,5-biphosphate (PI(4,5)P2) in a manner controlled by PKC and calcium/calmodulin. In neurons, manipulations of MARCKS concentration and membrane targeting strongly affect the numbers, shapes, and F-actin properties of dendritic spines, but the mechanisms remain unclear. Here, we tested the hypothesis that the effects of MARCKS on dendritic spine morphology are due to its capacity to regulate the availability of plasma membrane PI(4,5)P2. We observed that the concentration of free PI(4,5)P2 on the dendritic plasma membrane was inversely proportional to the concentration of MARCKS. Endogenous PI(4,5)P2 levels were increased or decreased, respectively, by acutely overexpressing either phosphatidylinositol-4-phosphate 5-kinase (PIP5K) or inositol polyphosphate 5-phosphatase (5ptase). PIP5K, like MARCKS depletion, induced severe spine shrinkage; 5ptase, like constitutively membrane-bound MARCKS, induced aberrant spine elongation. These phenotypes involved changes in actin properties driven by the F-actin severing protein cofilin. Collectively, these findings support a model in which neuronal activity regulates actin-dependent spine morphology through antagonistic interactions of MARCKS and PI(4,5)P2.
Figures



References
-
- Arbuzova A, Wang J, Murray D, Jacob J, Cafiso DS, McLaughlin S (1997). Kinetics of interaction of the myristoylated alanine-rich C kinase substrate, membranes, and calmodulin. J Biol Chem 272, 27167–27177. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

