Germline ablation achieved via CRISPR/Cas9 targeting of NANOS3 in bovine zygotes
- PMID: 38089499
- PMCID: PMC10711618
- DOI: 10.3389/fgeed.2023.1321243
Germline ablation achieved via CRISPR/Cas9 targeting of NANOS3 in bovine zygotes
Abstract
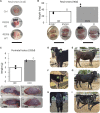
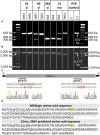
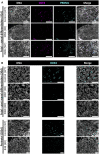
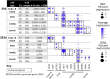
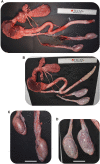
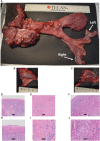
NANOS3 is expressed in migrating primordial germ cells (PGCs) to protect them from apoptosis, and it is known to be a critical factor for germline development of both sexes in several organisms. However, to date, live NANOS3 knockout (KO) cattle have not been reported, and the specific role of NANOS3 in male cattle, or bulls, remains unexplored. This study generated NANOS3 KO cattle via cytoplasmic microinjection of the CRISPR/Cas9 system in vitro produced bovine zygotes and evaluated the effect of NANOS3 elimination on bovine germline development, from fetal development through reproductive age. The co-injection of two selected guide RNA (gRNA)/Cas9 ribonucleoprotein complexes (i.e., dual gRNA approach) at 6 h post fertilization achieved a high NANOS3 KO rate in developing embryos. Subsequent embryo transfers resulted in a 31% (n = 8/26) pregnancy rate. A 75% (n = 6/8) total KO rate (i.e., 100% of alleles present contained complete loss-of-function mutations) was achieved with the dual gRNA editing approach. In NANOS3 KO fetal testes, PGCs were found to be completely eliminated by 41-day of fetal age. Importantly, despite the absence of germ cells, seminiferous tubule development was not impaired in NANOS3 KO bovine testes during fetal, perinatal, and adult stages. Moreover, a live, NANOS3 KO, germline-ablated bull was produced and at sexual maturity he exhibited normal libido, an anatomically normal reproductive tract, and intact somatic gonadal development and structure. Additionally, a live, NANOS3 KO, germline-ablated heifer was produced. However, it was evident that the absence of germ cells in NANOS3 KO cattle compromised the normalcy of ovarian development to a greater extent than it did testes development. The meat composition of NANOS3 KO cattle was unremarkable. Overall, this study demonstrated that the absence of NANOS3 in cattle leads to the specific deficiency of both male and female germ cells, suggesting the potential of NANOS3 KO cattle to act as hosts for donor-derived exogenous germ cell production in both sexes. These findings contribute to the understanding of NANOS3 function in cattle and have valuable implications for the development of novel breeding technologies using germline complementation in NANOS3 KO germline-ablated hosts.
Keywords: CRISPR/Cas9; NANOS3; bovine; embryo; gene editing; germline ablation.
Copyright © 2023 Mueller, McNabb, Owen, Hennig, Ledesma, Angove, Conley, Ross and Van Eenennaam.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

