A single-cell atlas of the peripheral immune response in patients with influenza A virus infection
- PMID: 38089584
- PMCID: PMC10711475
- DOI: 10.1016/j.isci.2023.108507
A single-cell atlas of the peripheral immune response in patients with influenza A virus infection
Abstract
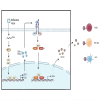
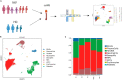
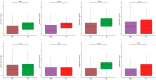
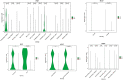
Influenza A virus (IAV) remains a pressing global health concern, yet our understanding of the specific nature and functional roles of certain circulating cell subsets in relation to this viral infection remains unclear. We performed single-cell RNA sequencing (scRNA-seq) on single-cell whole-blood (scWB) isolated from various populations using the Singleron Matrix platform. Our investigation showed a significant upregulation of the IFN-stimulated gene, IFN-α-inducible protein 27 (IFI27), in patients affected by IAV infection and further found that the heightened expression of IFI27 was primarily concentrated in specific immune cell populations, including monocytes and conventional dendritic cells (cDCs). Notably, we identified a specific subset of neutrophils, neutrophil_ISG15, which implicates interferon (IFN) signaling in IAV infection. Our findings provide a comprehensive understanding of the cellular subtypes and molecular characteristics of scWB across different populations with IAV infection.
Keywords: Immunology; Transcriptomics; Virology.
© 2023 The Authors.
Conflict of interest statement
Authors declare that they have no competing interests.
Figures






References
-
- World Health Organization Influenza (seasonal) 2023. http://www.who.int/mediacentre/factsheets/fs211/en/
-
- Poehling K.A., Edwards K.M., Weinberg G.A., Szilagyi P., Staat M.A., Iwane M.K., Bridges C.B., Grijalva C.G., Zhu Y., Bernstein D.I., et al. The underrecognized burden of influenza in young children. N. Engl. J. Med. 2006;355:31–40. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

