Identification of autophagy-related genes in osteoarthritis articular cartilage and their roles in immune infiltration
- PMID: 38090564
- PMCID: PMC10711085
- DOI: 10.3389/fimmu.2023.1263988
Identification of autophagy-related genes in osteoarthritis articular cartilage and their roles in immune infiltration
Abstract
Background: Autophagy plays a critical role in the progression of osteoarthritis (OA), mainly by regulating inflammatory and immune responses. However, the underlying mechanisms remain unclear. This study aimed to investigate the potential relevance of autophagy-related genes (ARGs) associated with infiltrating immune cells in OA.
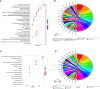
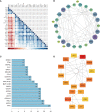
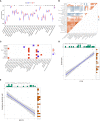
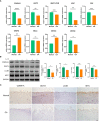
Methods: GSE114007, GSE169077, and ARGs were obtained from the Gene Expression Omnibus (GEO) database and the Human Autophagy database. R software was used to identify the differentially expressed autophagy-related genes (DEARGs) in OA. Functional enrichment and protein-protein interaction (PPI) analyses were performed to explore the role of DEARGs in OA cartilage, and then Cytoscape was utilized to screen hub ARGs. Single-sample gene set enrichment analysis (ssGSEA) was used to conduct immune infiltration analysis and evaluate the potential correlation of key ARGs and immune cell infiltration. Then, the expression levels of hub ARGs in OA were further verified by the GSE169077 and qRT-PCR. Finally, Western blotting and immunohistochemistry were used to validate the final hub ARGs.
Results: A total of 24 downregulated genes and five upregulated genes were identified, and these genes were enriched in autophagy, mitophagy, and inflammation-related pathways. The intersection results identified nine hub genes, namely, CDKN1A, DDIT3, FOS, VEGFA, RELA, MAP1LC3B, MYC, HSPA5, and HSPA8. GSE169077 and qRT-PCR validation results showed that only four genes, CDKN1A, DDT3, MAP1LC3B, and MYC, were consistent with the bioinformatics analysis results. Western blotting and immunohistochemical (IHC) showed that the expression of these four genes was significantly downregulated in the OA group, which is consistent with the qPCR results. Immune infiltration correlation analysis indicated that DDIT3 was negatively correlated with immature dendritic cells in OA, and FOS was positively correlated with eosinophils.
Conclusion: CDKN1A, DDIT3, MAP1LC3B, and MYC were identified as ARGs that were closely associated with immune infiltration in OA cartilage. Among them, DDIT3 showed a strong negative correlation with immature dendritic cells. This study found that the interaction between ARGs and immune cell infiltration may play a crucial role in the pathogenesis of OA; however, the specific interaction mechanism needs further research to be clarified. This study provides new insights to further understand the molecular mechanisms of immunity involved in the process of OA by autophagy.
Keywords: autophagy; bioinformatics analysis; cartilage; immune cell infiltration; osteoarthritis.
Copyright © 2023 Qin, Zhang, Wu, Ru, Zhong, Zhao and Lan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Gregori D, Giacovelli G, Minto C, Barbetta B, Gualtieri F, Azzolina D, et al. Association of pharmacological treatments with long-term pain control in patients with knee osteoarthritis: A systematic review and meta-analysis. JAMA (2018) 320(24):2564–79. doi: 10.1001/jama.2018.19319 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

