Killer instincts: natural killer cells as multifactorial cancer immunotherapy
- PMID: 38090565
- PMCID: PMC10715270
- DOI: 10.3389/fimmu.2023.1269614
Killer instincts: natural killer cells as multifactorial cancer immunotherapy
Abstract
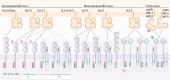
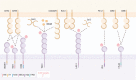
Natural killer (NK) cells integrate heterogeneous signals for activation and inhibition using germline-encoded receptors. These receptors are stochastically co-expressed, and their concurrent engagement and signaling can adjust the sensitivity of individual cells to putative targets. Against cancers, which mutate and evolve under therapeutic and immunologic pressure, the diversity for recognition provided by NK cells may be key to comprehensive cancer control. NK cells are already being trialled as adoptive cell therapy and targets for immunotherapeutic agents. However, strategies to leverage their naturally occurring diversity and agility have not yet been developed. In this review, we discuss the receptors and signaling pathways through which signals for activation or inhibition are generated in NK cells, focusing on their roles in cancer and potential as targets for immunotherapies. Finally, we consider the impacts of receptor co-expression and the potential to engage multiple pathways of NK cell reactivity to maximize the scope and strength of antitumor activities.
Keywords: cancer immunotherapy; innate lymphoid cells; killer immunoglobulin-like receptors (KIR); natural killer cells; signal integration.
Copyright © 2023 Nersesian, Carter, Lee, Westhaver and Boudreau.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

