Unraveling C-to-U RNA editing events from direct RNA sequencing
- PMID: 38090878
- PMCID: PMC10732634
- DOI: 10.1080/15476286.2023.2290843
Unraveling C-to-U RNA editing events from direct RNA sequencing
Abstract
In mammals, RNA editing events involve the conversion of adenosine (A) in inosine (I) by ADAR enzymes or the hydrolytic deamination of cytosine (C) in uracil (U) by the APOBEC family of enzymes, mostly APOBEC1. RNA editing has a plethora of biological functions, and its deregulation has been associated with various human disorders. While the large-scale detection of A-to-I is quite straightforward using the Illumina RNAseq technology, the identification of C-to-U events is a non-trivial task. This difficulty arises from the rarity of such events in eukaryotic genomes and the challenge of distinguishing them from background noise. Direct RNA sequencing by Oxford Nanopore Technology (ONT) permits the direct detection of Us on sequenced RNA reads. Surprisingly, using ONT reads from wild-type (WT) and APOBEC1-knock-out (KO) murine cell lines as well as in vitro synthesized RNA without any modification, we identified a systematic error affecting the accuracy of the Cs call, thereby leading to incorrect identifications of C-to-U events. To overcome this issue in direct RNA reads, here we introduce a novel machine learning strategy based on the isolation Forest (iForest) algorithm in which C-to-U editing events are considered as sequencing anomalies. Using in vitro synthesized and human ONT reads, our model optimizes the signal-to-noise ratio improving the detection of C-to-U editing sites with high accuracy, over 90% in all samples tested. Our results suggest that iForest, known for its rapid implementation and minimal memory requirements, is a promising tool to denoise ONT reads and reliably identify RNA modifications.
Keywords: C-to-U editing; Direct RNA sequencing; RNA editing; RNA modifications; iForest.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

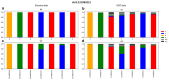

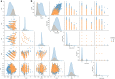
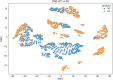
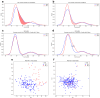
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
