Single-Cell Transcriptomics Reveals the Heterogeneity of the Immune Landscape of IDH-Wild-Type High-Grade Gliomas
- PMID: 38091354
- PMCID: PMC10835213
- DOI: 10.1158/2326-6066.CIR-23-0211
Single-Cell Transcriptomics Reveals the Heterogeneity of the Immune Landscape of IDH-Wild-Type High-Grade Gliomas
Abstract
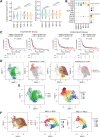
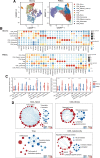
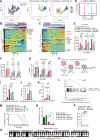
Isocitrate dehydrogenase (IDH)-wild-type (WT) high-grade gliomas, especially glioblastomas, are highly aggressive and have an immunosuppressive tumor microenvironment. Although tumor-infiltrating immune cells are known to play a critical role in glioma genesis, their heterogeneity and intercellular interactions remain poorly understood. In this study, we constructed a single-cell transcriptome landscape of immune cells from tumor tissue and matching peripheral blood mononuclear cells (PBMC) from IDH-WT high-grade glioma patients. Our analysis identified two subsets of tumor-associated macrophages (TAM) in tumors with the highest protumorigenesis signatures, highlighting their potential role in glioma progression. We also investigated the T-cell trajectory and identified the aryl hydrocarbon receptor (AHR) as a regulator of T-cell dysfunction, providing a potential target for glioma immunotherapy. We further demonstrated that knockout of AHR decreased chimeric antigen receptor (CAR) T-cell exhaustion and improved CAR T-cell antitumor efficacy both in vitro and in vivo. Finally, we explored intercellular communication mediated by ligand-receptor interactions within the tumor microenvironment and PBMCs and revealed the unique cellular interactions present in the tumor microenvironment. Taken together, our study provides a comprehensive immune landscape of IDH-WT high-grade gliomas and offers potential drug targets for glioma immunotherapy.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures



References
-
- Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol 2018;15:422–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

