Myeloid/natural killer (NK) cell precursor acute leukemia as a distinct leukemia type
- PMID: 38091391
- PMCID: PMC10848711
- DOI: 10.1126/sciadv.adj4407
Myeloid/natural killer (NK) cell precursor acute leukemia as a distinct leukemia type
Abstract
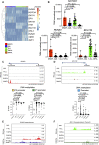
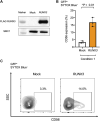
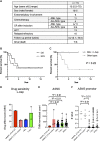
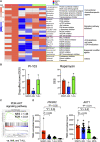
Myeloid/natural killer (NK) cell precursor acute leukemia (MNKPL) has been described on the basis of its unique immunophenotype and clinical phenotype. However, there is no consensus on the characteristics for identifying this disease type because of its rarity and lack of defined distinctive molecular characteristics. In this study, multiomics analysis revealed that MNKPL is distinct from acute myeloid leukemia, T cell acute lymphoblastic leukemia, and mixed-phenotype acute leukemia (MPAL), and NOTCH1 and RUNX3 activation and BCL11B down-regulation are hallmarks of MNKPL. Although NK cells have been classically considered to be lymphoid lineage-derived, the results of our single-cell analysis using MNKPL cells suggest that NK cells and myeloid cells share common progenitor cells. Treatment outcomes for MNKPL are unsatisfactory, even when hematopoietic cell transplantation is performed. Multiomics analysis and in vitro drug sensitivity assays revealed increased sensitivity to l-asparaginase and reduced levels of asparagine synthetase (ASNS), supporting the clinically observed effectiveness of l-asparaginase.
Figures



References
-
- R. Suzuki, K. Yamamoto, M. Seto, Y. Kagami, M. Ogura, Y. Yatabe, T. Suchi, Y. Kodera, Y. Morishima, T. Takahashi, H. Saito, R. Ueda, S. Nakamura, CD7+ and CD56+ myeloid/natural killer cell precursor acute leukemia: A distinct hematolymphoid disease entity. Blood 90, 2417–2428 (1997). - PubMed
-
- Y. Noguchi, D. Tomizawa, H. Hiroki, S. Miyamoto, M. Tezuka, R. Miyawaki, M. Tanaka-Kubota, T. Okano, C. Kobayashi, N. Mitsuiki, Y. Aoki, K. Imai, M. Kajiwara, H. Kanegane, T. Morio, M. Takagi, Hematopoietic cell transplantation for myeloid/NK cell precursor acute leukemia in second remission. Clin. Case Rep. 6, 1023–1028 (2018). - PMC - PubMed
-
- Y. Ma, B. Chen, X. Xu, G. Lin, Myeloid/natural killer cell precursor acute leukemia with multiple subcutaneous nodules as the initial presentation: A case report and literature review. Int. J. Hematol. 90, 243–247 (2009). - PubMed
-
- X. Liang, D. K. Graham, Natural killer cell neoplasms. Cancer 112, 1425–1436 (2008). - PubMed
-
- A. A. Scott, D. R. Head, K. J. Kopecky, F. R. Appelbaum, K. S. Theil, M. R. Grever, I. M. Chen, M. H. Whittaker, B. B. Griffith, J. D. Licht, S. Waxman, M. M. Whalen, A. D. Bankhurst, L. C. Richter, T. M. Grogan, C. L. Willman, HLA-DR, CD33+, CD56+, CD16- myeloid/natural killer cell acute leukemia: A previously unrecognized form of acute leukemia potentially misdiagnosed as French-American-British acute myeloid leukemia-M3. Blood 84, 244–255 (1994). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

