Circulating senescent myeloid cells infiltrate the brain and cause neurodegeneration in histiocytic disorders
- PMID: 38091952
- PMCID: PMC11587932
- DOI: 10.1016/j.immuni.2023.11.011
Circulating senescent myeloid cells infiltrate the brain and cause neurodegeneration in histiocytic disorders
Abstract
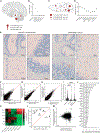
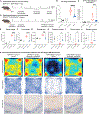
Neurodegenerative diseases (ND) are characterized by progressive loss of neuronal function. Mechanisms of ND pathogenesis are incompletely understood, hampering the development of effective therapies. Langerhans cell histiocytosis (LCH) is an inflammatory neoplastic disorder caused by hematopoietic progenitors expressing mitogen-activated protein kinase (MAPK)-activating mutations that differentiate into senescent myeloid cells that drive lesion formation. Some individuals with LCH subsequently develop progressive and incurable neurodegeneration (LCH-ND). Here, we showed that LCH-ND was caused by myeloid cells that were clonal with peripheral LCH cells. Circulating BRAFV600E+ myeloid cells caused the breakdown of the blood-brain barrier (BBB), enhancing migration into the brain parenchyma where they differentiated into senescent, inflammatory CD11a+ macrophages that accumulated in the brainstem and cerebellum. Blocking MAPK activity and senescence programs reduced peripheral inflammation, brain parenchymal infiltration, neuroinflammation, neuronal damage and improved neurological outcome in preclinical LCH-ND. MAPK activation and senescence programs in circulating myeloid cells represent targetable mechanisms of LCH-ND.
Keywords: LCH-ND; Langerhans cell histiocytosis; blood-brain barrier; histiocytic disorders; neurodegeneration; neuroinflammation.
Copyright © 2023. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Update of
-
Circulating senescent myeloid cells drive blood brain barrier breakdown and neurodegeneration.bioRxiv [Preprint]. 2023 Oct 11:2023.10.10.561744. doi: 10.1101/2023.10.10.561744. bioRxiv. 2023. Update in: Immunity. 2023 Dec 12;56(12):2790-2802.e6. doi: 10.1016/j.immuni.2023.11.011. PMID: 37873371 Free PMC article. Updated. Preprint.
Comment in
-
A maelstrom of migrating monocytes drives neurodegeneration.Immunity. 2023 Dec 12;56(12):2677-2678. doi: 10.1016/j.immuni.2023.11.014. Immunity. 2023. PMID: 38091948
References
-
- Allen CE, Flores R, Rauch R, Dauser R, Murray JC, Puccetti D, Hsu DA, Sondel P, Hetherington M, Goldman S, and McClain KL (2010). Neurodegenerative central nervous system Langerhans cell histiocytosis and coincident hydrocephalus treated with vincristine/cytosine arabinoside. Pediatr. Blood Cancer 54, 416–423. - PMC - PubMed
-
- Héritier S, Barkaoui MA, Miron J, Thomas C, Moshous D, Lambilliotte A, Mazingue F, Kebaili K, Jeziorski E, Plat G, et al. (2018). Incidence and risk factors for clinical neurodegenerative Langerhans cell histiocytosis: a longitudinal cohort study. Br. J. Haematol 183, 608–617. - PubMed
-
- Yeh EA, Greenberg J, Abla O, Longoni G, Diamond E, Hermiston M, Tran B, Rodriguez-Galindo C, Allen CE, and McClain KL; North American Consortium for Histiocytosis (2018). Evaluation and treatment of Langerhans cell histiocytosis patients with central nervous system abnormalities: Current views and new vistas. Pediatr. Blood Cancer 65, e26784. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

