iPSC-derived hypoimmunogenic tissue resident memory T cells mediate robust anti-tumor activity against cervical cancer
- PMID: 38091985
- PMCID: PMC10772465
- DOI: 10.1016/j.xcrm.2023.101327
iPSC-derived hypoimmunogenic tissue resident memory T cells mediate robust anti-tumor activity against cervical cancer
Abstract
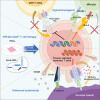
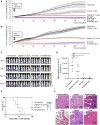
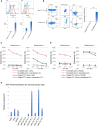
Functionally rejuvenated human papilloma virus-specific cytotoxic T lymphocytes (HPV-rejTs) generated from induced pluripotent stem cells robustly suppress cervical cancer. However, autologous rejT generation is time consuming, leading to difficulty in treating patients with advanced cancer. Although use of allogeneic HPV-rejTs can obviate this, the major obstacle is rejection by the patient immune system. To overcome this, we develop HLA-A24&-E dual integrated HPV-rejTs after erasing HLA class I antigens. These rejTs effectively suppress recipient immune rejection while maintaining more robust cytotoxicity than original cytotoxic T lymphocytes. Single-cell RNA sequencing performed to gain deeper insights reveal that HPV-rejTs are highly enriched with tissue resident memory T cells, which enhance cytotoxicity against cervical cancer through TGFβR signaling, with increased CD103 expression. Genes associated with the immunological synapse also are upregulated, suggesting that these features promote stronger activation of T cell receptor (TCR) and increased TCR-mediated target cell death. We believe that our work will contribute to feasible "off-the-shelf" T cell therapy with robust anti-cervical cancer effects.
Keywords: CRISPR-Cas9 scarless gene editing; HLA class I-edited iPSCs; cervical cancer; human papilloma virus-specific CTLs; hypoimmunogenic iPSC-derived CTLs; rejuvenated CTLs; single-cell RNA sequencing; tissue resident memory T cells; two-step gene editing; “off-the-shelf” CTL therapy.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests K.I. is currently chief executive officer of Bayspair Inc. and an inventor of US11692202B2. M.P. serves on the Scientific Advisory Board and has equity in Allogene Tx, serves on the Board of Directors and has equity in Graphite Biologics, has equity in CRISPR Therapeutics, and serves as a consultant to Bayspair. H.N. is a co-founder of Century Therapeutics.
Figures




References
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. - PubMed
-
- de Villiers E.M. Cross-roads in the classification of papillomaviruses. Virology. 2013;445:2–10. - PubMed
-
- Parkin D.M., Bray F. Chapter 2: The burden of HPV-related cancers. Vaccine. 2006;24:11–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

