Combined usage of serodiagnosis and O antigen typing to isolate Shiga toxin-producing Escherichia coli O76:H7 from a hemolytic uremic syndrome case and genomic insights from the isolate
- PMID: 38092668
- PMCID: PMC10790564
- DOI: 10.1128/spectrum.02355-23
Combined usage of serodiagnosis and O antigen typing to isolate Shiga toxin-producing Escherichia coli O76:H7 from a hemolytic uremic syndrome case and genomic insights from the isolate
Abstract
Hemolytic uremic syndrome (HUS) is a life-threatening disease caused by Shiga toxin-producing Escherichia coli (STEC) infection. The treatment approaches for STEC-mediated typical HUS and atypical HUS differ, underscoring the importance of rapid and accurate diagnosis. However, specific detection methods for STECs other than major serogroups, such as O157, O26, and O111, are limited. This study focuses on the utility of PCR-based O-serotyping, serum agglutination tests utilizing antibodies against the identified Og type, and isolation techniques employing antibody-conjugated immunomagnetic beads for STEC isolation. By employing these methods, we successfully isolated a STEC strain of a minor serotype, O76:H7, from a HUS patient.
Keywords: Shiga toxin-producing Escherichia coli; genome analysis; hemolytic uremic syndrome; immunomagnetic separation; immunoserology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


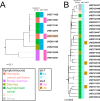

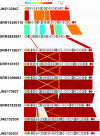
References
-
- Infectious Agents Surveillance Report . 2020. Enterohemorrhagic Escherichia coli (EHEC) infection as of March 2020 in Japan. IASR 41:65–66. doi: 10.7883/yoken.JJID.2008.58 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

