DPPIV+ fibro-adipogenic progenitors form the niche of adult skeletal muscle self-renewing resident macrophages
- PMID: 38092736
- PMCID: PMC10719395
- DOI: 10.1038/s41467-023-43579-3
DPPIV+ fibro-adipogenic progenitors form the niche of adult skeletal muscle self-renewing resident macrophages
Abstract
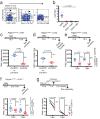
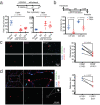
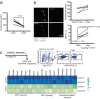
Adult tissue-resident macrophages (RMs) are either maintained by blood monocytes or through self-renewal. While the presence of a nurturing niche is likely crucial to support the survival and function of self-renewing RMs, evidence regarding its nature is limited. Here, we identify fibro-adipogenic progenitors (FAPs) as the main source of colony-stimulating factor 1 (CSF1) in resting skeletal muscle. Using parabiosis in combination with FAP-deficient transgenic mice (PdgfrαCreERT2 × DTA) or mice lacking FAP-derived CSF1 (PdgfrαCreERT2 × Csf1flox/null), we show that local CSF1 from FAPs is required for the survival of both TIM4- monocyte-derived and TIM4+ self-renewing RMs in adult skeletal muscle. The spatial distribution and number of TIM4+ RMs coincide with those of dipeptidyl peptidase IV (DPPIV)+ FAPs, suggesting their role as CSF1-producing niche cells for self-renewing RMs. This finding identifies opportunities to precisely manipulate the function of self-renewing RMs in situ to further unravel their role in health and disease.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

