COVID-19 and influenza infections mediate distinct pulmonary cellular and transcriptomic changes
- PMID: 38092883
- PMCID: PMC10719262
- DOI: 10.1038/s42003-023-05626-z
COVID-19 and influenza infections mediate distinct pulmonary cellular and transcriptomic changes
Abstract
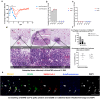
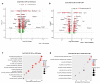
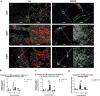
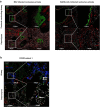
SARS-CoV-2 infection can cause persistent respiratory sequelae. However, the underlying mechanisms remain unclear. Here we report that sub-lethally infected K18-human ACE2 mice show patchy pneumonia associated with histiocytic inflammation and collagen deposition at 21 and 45 days post infection (DPI). Transcriptomic analyses revealed that compared to influenza-infected mice, SARS-CoV-2-infected mice had reduced interferon-gamma/alpha responses at 4 DPI and failed to induce keratin 5 (Krt5) at 6 DPI in lung, a marker of nascent pulmonary progenitor cells. Histologically, influenza- but not SARS-CoV-2-infected mice showed extensive Krt5+ "pods" structure co-stained with stem cell markers Trp63/NGFR proliferated in the pulmonary consolidation area at both 7 and 14 DPI, with regression at 21 DPI. These Krt5+ "pods" structures were not observed in the lungs of SARS-CoV-2-infected humans or nonhuman primates. These results suggest that SARS-CoV-2 infection fails to induce nascent Krt5+ cell proliferation in consolidated regions, leading to incomplete repair of the injured lung.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

