Molecularly defined and spatially resolved cell atlas of the whole mouse brain
- PMID: 38092912
- PMCID: PMC10719103
- DOI: 10.1038/s41586-023-06808-9
Molecularly defined and spatially resolved cell atlas of the whole mouse brain
Abstract
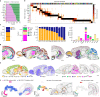
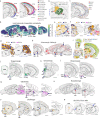
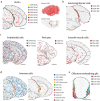
In mammalian brains, millions to billions of cells form complex interaction networks to enable a wide range of functions. The enormous diversity and intricate organization of cells have impeded our understanding of the molecular and cellular basis of brain function. Recent advances in spatially resolved single-cell transcriptomics have enabled systematic mapping of the spatial organization of molecularly defined cell types in complex tissues1-3, including several brain regions (for example, refs. 1-11). However, a comprehensive cell atlas of the whole brain is still missing. Here we imaged a panel of more than 1,100 genes in approximately 10 million cells across the entire adult mouse brains using multiplexed error-robust fluorescence in situ hybridization12 and performed spatially resolved, single-cell expression profiling at the whole-transcriptome scale by integrating multiplexed error-robust fluorescence in situ hybridization and single-cell RNA sequencing data. Using this approach, we generated a comprehensive cell atlas of more than 5,000 transcriptionally distinct cell clusters, belonging to more than 300 major cell types, in the whole mouse brain with high molecular and spatial resolution. Registration of this atlas to the mouse brain common coordinate framework allowed systematic quantifications of the cell-type composition and organization in individual brain regions. We further identified spatial modules characterized by distinct cell-type compositions and spatial gradients featuring gradual changes of cells. Finally, this high-resolution spatial map of cells, each with a transcriptome-wide expression profile, allowed us to infer cell-type-specific interactions between hundreds of cell-type pairs and predict molecular (ligand-receptor) basis and functional implications of these cell-cell interactions. These results provide rich insights into the molecular and cellular architecture of the brain and a foundation for functional investigations of neural circuits and their dysfunction in health and disease.
© 2023. The Author(s).
Conflict of interest statement
X.Z. is a co-founder and consultant of Vizgen. H.Z. is on the scientific advisory board of MapLight Therapeutics, Inc.
Figures












Update of
-
A molecularly defined and spatially resolved cell atlas of the whole mouse brain.bioRxiv [Preprint]. 2023 Mar 7:2023.03.06.531348. doi: 10.1101/2023.03.06.531348. bioRxiv. 2023. Update in: Nature. 2023 Dec;624(7991):343-354. doi: 10.1038/s41586-023-06808-9. PMID: 36945367 Free PMC article. Updated. Preprint.
Similar articles
-
A molecularly defined and spatially resolved cell atlas of the whole mouse brain.bioRxiv [Preprint]. 2023 Mar 7:2023.03.06.531348. doi: 10.1101/2023.03.06.531348. bioRxiv. 2023. Update in: Nature. 2023 Dec;624(7991):343-354. doi: 10.1038/s41586-023-06808-9. PMID: 36945367 Free PMC article. Updated. Preprint.
-
Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH.Nature. 2021 Oct;598(7879):137-143. doi: 10.1038/s41586-021-03705-x. Epub 2021 Oct 6. Nature. 2021. PMID: 34616063 Free PMC article.
-
Computational solutions for spatial transcriptomics.Comput Struct Biotechnol J. 2022 Sep 1;20:4870-4884. doi: 10.1016/j.csbj.2022.08.043. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36147664 Free PMC article. Review.
-
Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression.Proc Natl Acad Sci U S A. 2019 Sep 24;116(39):19490-19499. doi: 10.1073/pnas.1912459116. Epub 2019 Sep 9. Proc Natl Acad Sci U S A. 2019. PMID: 31501331 Free PMC article.
-
Uncovering an Organ's Molecular Architecture at Single-Cell Resolution by Spatially Resolved Transcriptomics.Trends Biotechnol. 2021 Jan;39(1):43-58. doi: 10.1016/j.tibtech.2020.05.006. Epub 2020 Jun 3. Trends Biotechnol. 2021. PMID: 32505359 Review.
Cited by
-
Optics-free reconstruction of 2D images via DNA barcode proximity graphs.bioRxiv [Preprint]. 2024 Aug 8:2024.08.06.606834. doi: 10.1101/2024.08.06.606834. bioRxiv. 2024. PMID: 39149271 Free PMC article. Preprint.
-
Shaping the olfactory map: cell type-specific activity patterns guide circuit formation.Front Neural Circuits. 2024 May 27;18:1409680. doi: 10.3389/fncir.2024.1409680. eCollection 2024. Front Neural Circuits. 2024. PMID: 38860141 Free PMC article. Review.
-
Network analysis of α-synuclein pathology progression reveals p21-activated kinases as regulators of vulnerability.bioRxiv [Preprint]. 2024 Oct 22:2024.10.22.619411. doi: 10.1101/2024.10.22.619411. bioRxiv. 2024. PMID: 39484617 Free PMC article. Preprint.
-
SC2Spa: a deep learning based approach to map transcriptome to spatial origins at cellular resolution.BMC Bioinformatics. 2025 Jun 2;26(1):148. doi: 10.1186/s12859-025-06173-6. BMC Bioinformatics. 2025. PMID: 40457183 Free PMC article.
-
Opportunities and challenges of single-cell and spatially resolved genomics methods for neuroscience discovery.Nat Neurosci. 2024 Dec;27(12):2292-2309. doi: 10.1038/s41593-024-01806-0. Epub 2024 Dec 3. Nat Neurosci. 2024. PMID: 39627587 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

