In vitro production of cat-restricted Toxoplasma pre-sexual stages
- PMID: 38093015
- PMCID: PMC10781626
- DOI: 10.1038/s41586-023-06821-y
In vitro production of cat-restricted Toxoplasma pre-sexual stages
Abstract
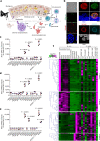
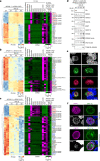
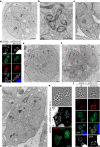
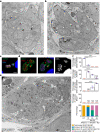
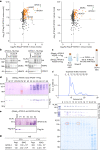
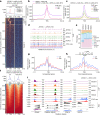
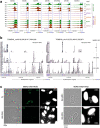
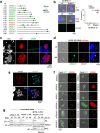
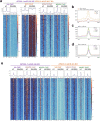
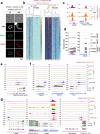
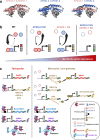
Sexual reproduction of Toxoplasma gondii, confined to the felid gut, remains largely uncharted owing to ethical concerns regarding the use of cats as model organisms. Chromatin modifiers dictate the developmental fate of the parasite during its multistage life cycle, but their targeting to stage-specific cistromes is poorly described1,2. Here we found that the transcription factors AP2XII-1 and AP2XI-2 operate during the tachyzoite stage, a hallmark of acute toxoplasmosis, to silence genes necessary for merozoites, a developmental stage critical for subsequent sexual commitment and transmission to the next host, including humans. Their conditional and simultaneous depletion leads to a marked change in the transcriptional program, promoting a full transition from tachyzoites to merozoites. These in vitro-cultured pre-gametes have unique protein markers and undergo typical asexual endopolygenic division cycles. In tachyzoites, AP2XII-1 and AP2XI-2 bind DNA as heterodimers at merozoite promoters and recruit MORC and HDAC3 (ref. 1), thereby limiting chromatin accessibility and transcription. Consequently, the commitment to merogony stems from a profound epigenetic rewiring orchestrated by AP2XII-1 and AP2XI-2. Successful production of merozoites in vitro paves the way for future studies on Toxoplasma sexual development without the need for cat infections and holds promise for the development of therapies to prevent parasite transmission.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
In vitro production of cat-restricted Toxoplasma pre-sexual stages by epigenetic reprogramming.bioRxiv [Preprint]. 2023 Jan 17:2023.01.16.524187. doi: 10.1101/2023.01.16.524187. bioRxiv. 2023. Update in: Nature. 2024 Jan;625(7994):366-376. doi: 10.1038/s41586-023-06821-y. PMID: 36711883 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

