Bioactive glycans in a microbiome-directed food for children with malnutrition
- PMID: 38093016
- PMCID: PMC10764277
- DOI: 10.1038/s41586-023-06838-3
Bioactive glycans in a microbiome-directed food for children with malnutrition
Abstract
Evidence is accumulating that perturbed postnatal development of the gut microbiome contributes to childhood malnutrition1-4. Here we analyse biospecimens from a randomized, controlled trial of a microbiome-directed complementary food (MDCF-2) that produced superior rates of weight gain compared with a calorically more dense conventional ready-to-use supplementary food in 12-18-month-old Bangladeshi children with moderate acute malnutrition4. We reconstructed 1,000 bacterial genomes (metagenome-assembled genomes (MAGs)) from the faecal microbiomes of trial participants, identified 75 MAGs of which the abundances were positively associated with ponderal growth (change in weight-for-length Z score (WLZ)), characterized changes in MAG gene expression as a function of treatment type and WLZ response, and quantified carbohydrate structures in MDCF-2 and faeces. The results reveal that two Prevotella copri MAGs that are positively associated with WLZ are the principal contributors to MDCF-2-induced expression of metabolic pathways involved in utilizing the component glycans of MDCF-2. The predicted specificities of carbohydrate-active enzymes expressed by their polysaccharide-utilization loci are correlated with (1) the in vitro growth of Bangladeshi P. copri strains, possessing varying degrees of polysaccharide-utilization loci and genomic conservation with these MAGs, in defined medium containing different purified glycans representative of those in MDCF-2, and (2) the levels of faecal carbohydrate structures in the trial participants. These associations suggest that identifying bioactive glycan structures in MDCFs metabolized by growth-associated bacterial taxa will help to guide recommendations about their use in children with acute malnutrition and enable the development of additional formulations.
© 2023. The Author(s).
Conflict of interest statement
A.L.O. and D.A.R. are co-founders of Phenobiome, a company pursuing development of computational tools for predictive phenotype profiling of microbial communities. C.B.L. is a co-founder of Infinant Health, interVenn Bio and BCD Bioscience—companies involved in the characterization of glycans and developing carbohydrate applications for human health. A joint patent application between Washington University in St Louis and icddr,b has been filed, entitled “Synbiotic combination of selected strains of
Figures

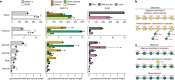













Update of
-
Bioactive glycans in a microbiome-directed food for malnourished children.medRxiv [Preprint]. 2023 Aug 18:2023.08.14.23293998. doi: 10.1101/2023.08.14.23293998. medRxiv. 2023. Update in: Nature. 2024 Jan;625(7993):157-165. doi: 10.1038/s41586-023-06838-3. PMID: 37645824 Free PMC article. Updated. Preprint.
References
-
- Levels and Trends in Child Malnutrition: UNICEF/WHO/The World Bank Group Joint Child Malnutrition Estimates: Key Findings of the 2021 Edition (WHO, 2021).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

