Structure-based virtual screening and in vitro validation of inhibitors of cyclic dinucleotide phosphodiesterases ENPP1 and CdnP
- PMID: 38095464
- PMCID: PMC10783014
- DOI: 10.1128/spectrum.02012-23
Structure-based virtual screening and in vitro validation of inhibitors of cyclic dinucleotide phosphodiesterases ENPP1 and CdnP
Abstract
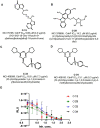
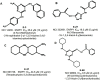
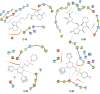
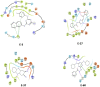
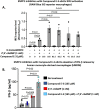
In this paper, we describe novel inhibitors of cyclic dinucleotide phosphodiesterase enzymes from Mycobacterium tuberculosis (M.tb) (CdnP) and mammals (ENPP1). The phosphodiesterase enzymes hydrolyze cyclic dinucleotides, such as 2',3'-cyclic GMP-AMP and c-di-AMP, which are stimulator of interferon gene (STING) agonists. By blocking the hydrolysis of STING agonists, the cyclic GMP-AMP synthase (cGAS)-STING-IRF3 pathway is potentiated. There is strong evidence in tuberculosis and in cancer biology that potentiation of the cGAS-STING-IRF3 pathway leads to improved M.tb clearance and also improved antitumor responses in cancer. In addition to the identification of novel inhibitors and their biochemical characterization, we provide proof-of-concept evidence that our E-3 inhibitor potentiates the cGAS-STING-IRF3 pathway in both macrophage cell lines and also in primary human monocyte-derived macrophages.
Keywords: Mycobacterium tuberculosis; cyclic dinucleotide phosphodiesterase; host-directed therapy; immune evasion.
Conflict of interest statement
W.R.B. is a co-founder of OncoSTING, LLC, which has licensed Johns Hopkins technology involving BCG strains that overexpress STING agonists.
Figures

Similar articles
-
Discovery of natural CdnP inhibitors through structure-based virtual screening and molecular dynamics simulations.Microbiol Spectr. 2025 Jun 3;13(6):e0325824. doi: 10.1128/spectrum.03258-24. Epub 2025 Apr 30. Microbiol Spectr. 2025. PMID: 40304517 Free PMC article.
-
Identification of a Mycobacterium tuberculosis Cyclic Dinucleotide Phosphodiesterase Inhibitor.ACS Infect Dis. 2021 Feb 12;7(2):309-317. doi: 10.1021/acsinfecdis.0c00444. Epub 2021 Jan 25. ACS Infect Dis. 2021. PMID: 33492938
-
Tumor Exosomal ENPP1 Hydrolyzes cGAMP to Inhibit cGAS-STING Signaling.Adv Sci (Weinh). 2024 May;11(20):e2308131. doi: 10.1002/advs.202308131. Epub 2024 Mar 18. Adv Sci (Weinh). 2024. PMID: 38498770 Free PMC article.
-
Ecto-nucleotide pyrophosphatase/phosphodiesterase 1 inhibitors: Research progress and prospects.Eur J Med Chem. 2024 Mar 5;267:116211. doi: 10.1016/j.ejmech.2024.116211. Epub 2024 Feb 10. Eur J Med Chem. 2024. PMID: 38359537 Review.
-
ENPP1, an Old Enzyme with New Functions, and Small Molecule Inhibitors-A STING in the Tale of ENPP1.Molecules. 2019 Nov 19;24(22):4192. doi: 10.3390/molecules24224192. Molecules. 2019. PMID: 31752288 Free PMC article. Review.
Cited by
-
Discovery of natural CdnP inhibitors through structure-based virtual screening and molecular dynamics simulations.Microbiol Spectr. 2025 Jun 3;13(6):e0325824. doi: 10.1128/spectrum.03258-24. Epub 2025 Apr 30. Microbiol Spectr. 2025. PMID: 40304517 Free PMC article.
-
Inhibitors of Cyclic Dinucleotide Phosphodiesterases and Cyclic Oligonucleotide Ring Nucleases as Potential Drugs for Various Diseases.Cells. 2025 Apr 30;14(9):663. doi: 10.3390/cells14090663. Cells. 2025. PMID: 40358186 Free PMC article. Review.
-
Role of ENPP1 in cancer pathogenesis: Mechanisms and clinical implications (Review).Oncol Lett. 2024 Oct 3;28(6):590. doi: 10.3892/ol.2024.14722. eCollection 2024 Dec. Oncol Lett. 2024. PMID: 39411204 Free PMC article. Review.
-
Recent advances in microbially derived chlorinated antiparasitic compounds.Mol Divers. 2024 Oct 30. doi: 10.1007/s11030-024-11018-0. Online ahead of print. Mol Divers. 2024. PMID: 39476277 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

