Uncovering a unique pathogenic mechanism of SARS-CoV-2 omicron variant: selective induction of cellular senescence
- PMID: 38095608
- PMCID: PMC10756098
- DOI: 10.18632/aging.205297
Uncovering a unique pathogenic mechanism of SARS-CoV-2 omicron variant: selective induction of cellular senescence
Abstract
Background: SARS-CoV-2 variants are constantly emerging with a variety of changes in the conformation of the spike protein, resulting in alterations of virus entry mechanisms. Solely omicron variants use the endosomal clathrin-mediated entry. Here, we investigate the influence of defined altered spike formations to study their impact on premature cellular senescence.
Methods: In our study, in vitro infections of SARS-CoV-2 variants delta (B.1.617.2) and omicron (B.1.1.529) were analyzed by using human primary small alveolar epithelial cells and human ex vivo lung slices. We confirmed cellular senescence in human lungs of COVID-19 patients. Hence, global gene expression patterns of infected human primary alveolar epithelial cells were identified via mRNA sequencing.
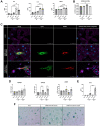
Results: Solely omicron variants of SARS-CoV-2 influenced the expression of cell cycle genes, highlighted by an increased p21 expression in human primary lung cells and human ex vivo lungs. Additionally, an upregulated senescence-associated secretory phenotype (SASP) was detected. Transcriptomic data indicate an increased gene expression of p16, and p38 in omicron-infected lung cells.
Conclusions: Significant changes due to different SARS-CoV-2 infections in human primary alveolar epithelial cells with an overall impact on premature aging could be identified. A substantially different cellular response with an upregulation of cell cycle, inflammation- and integrin-associated pathways in omicron infected cells indicates premature cellular senescence.
Keywords: SARS-CoV-2; cellular senescence; lung airway cells; variant of concern.
Conflict of interest statement
Figures




References
-
- Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Peacock SJ, Robertson DL, and COVID-19 Genomics UK (COG-UK) Consortium. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021; 19:409–24. 10.1038/s41579-021-00573-0 - DOI - PMC - PubMed
-
- Meng B, Abdullahi A, Ferreira IAT, Goonawardane N, Saito A, Kimura I, Yamasoba D, Gerber PP, Fatihi S, Rathore S, Zepeda SK, Papa G, Kemp SA, et al., and CITIID-NIHR BioResource COVID-19 Collaboration, and Genotype to Phenotype Japan (G2P-Japan) Consortium, and Ecuador-COVID19 Consortium. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022; 603:706–14. 10.1038/s41586-022-04474-x - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

