Conjoint research of WGCNA, single-cell transcriptome and structural biology reveals the potential targets of IDD development and treatment and JAK3 involvement
- PMID: 38095643
- PMCID: PMC10781489
- DOI: 10.18632/aging.205289
Conjoint research of WGCNA, single-cell transcriptome and structural biology reveals the potential targets of IDD development and treatment and JAK3 involvement
Abstract
Objectives: This study conducted integrated analysis of bulk RNA sequencing, single-cell RNA sequencing and Weighted Gene Co-expression Network Analysis (WGCNA), to comprehensively decode the most essential genes of intervertebral disc degeneration (IDD); then mainly focused on the JAK3 macromolecule to identify natural compounds to provide more candidate drug options in alleviating IDD.
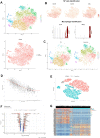
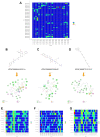
Methods: In the first part, we performed single-cell transcriptome analysis and WGCNA workflow to delineate the most pivotal genes of IDD. Then series of structural biology approaches and high-throughput virtual screening techniques were performed to discover potential compounds targeting JAK-STAT signaling pathway, such as Libdock, ADMET, precise molecular docking algorithm and in-vivo drug stability assessment.
Results: Totally 4 hub genes were determined in the development of IDD, namely VEGFA, MMP3, TNFSF11, and TIMP3, respectively. Then, 3 novel natural materials, ZINC000014952116, ZINC000003938642 and ZINC000072131515, were determined as potential compounds, with less toxicities and moderate ADME characteristics. In-vivo drug stability assessment suggested that these drugs could interact with JAK3, and their ligand-JAK3 complexes maintained the homeostasis in-vivo, which acted as regulatory role to JAK3 protein. Among them, ZINC000072131515, also known as Menaquinone, demonstrated significant protective roles to alleviate the progression of IDD in vitro, which proved the nutritional therapy in alleviating IDD.
Conclusions: This study reported the essential genes in the development of IDD, and also the roles of Menaquinone to ameliorate IDD through inhibiting JAK3 protein. This study also provided more options and resources on JAK3 targeted screening, which may further expand the drug resources in the pharmaceutical market.
Keywords: Janus kinase family; high-throughput virtual screening; intervertebral disc degeneration; single-cell transcriptome; weighted gene co-expression network analysis.
Conflict of interest statement
Figures








References
-
- Lewis RA, Williams NH, Sutton AJ, Burton K, Din NU, Matar HE, Hendry M, Phillips CJ, Nafees S, Fitzsimmons D, Rickard I, Wilkinson C. Comparative clinical effectiveness of management strategies for sciatica: systematic review and network meta-analyses. Spine J. 2015; 15:1461–77. 10.1016/j.spinee.2013.08.049 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

