Synonymous codon usage regulates translation initiation
- PMID: 38096059
- PMCID: PMC10790568
- DOI: 10.1016/j.celrep.2023.113413
Synonymous codon usage regulates translation initiation
Abstract
Nonoptimal synonymous codons repress gene expression, but the underlying mechanisms are poorly understood. We and others have previously shown that nonoptimal codons slow translation elongation speeds and thereby trigger messenger RNA (mRNA) degradation. Nevertheless, transcript levels are often insufficient to explain protein levels, suggesting additional mechanisms by which codon usage regulates gene expression. Using reporters in human and Drosophila cells, we find that transcript levels account for less than half of the variation in protein abundance due to codon usage. This discrepancy is explained by translational differences whereby nonoptimal codons repress translation initiation. Nonoptimal transcripts are also less bound by the translation initiation factors eIF4E and eIF4G1, providing a mechanistic explanation for their reduced initiation rates. Importantly, translational repression can occur without mRNA decay and deadenylation, and it does not depend on the known nonoptimality sensor, CNOT3. Our results reveal a potent mechanism of regulation by codon usage where nonoptimal codons repress further rounds of translation.
Keywords: CNOT3; CP: Molecular biology; codon optimality; deadenylation; eIF4E; gene regulation; mRNA decay; ribosome; translation initiation; translational repression.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests O.S.R. is a member of the Molecular Cell advisory board and the Cell Reports advisory board.
Figures


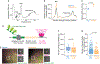



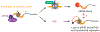
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

