Genetically corrected RAG2-SCID human hematopoietic stem cells restore V(D)J-recombinase and rescue lymphoid deficiency
- PMID: 38096800
- PMCID: PMC11006817
- DOI: 10.1182/bloodadvances.2023011766
Genetically corrected RAG2-SCID human hematopoietic stem cells restore V(D)J-recombinase and rescue lymphoid deficiency
Abstract
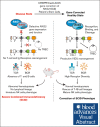
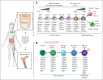
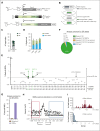
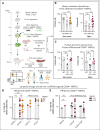
Recombination-activating genes (RAG1 and RAG2) are critical for lymphoid cell development and function by initiating the variable (V), diversity (D), and joining (J) (V(D)J)-recombination process to generate polyclonal lymphocytes with broad antigen specificity. The clinical manifestations of defective RAG1/2 genes range from immune dysregulation to severe combined immunodeficiencies (SCIDs), causing life-threatening infections and death early in life without hematopoietic cell transplantation (HCT). Despite improvements, haploidentical HCT without myeloablative conditioning carries a high risk of graft failure and incomplete immune reconstitution. The RAG complex is only expressed during the G0-G1 phase of the cell cycle in the early stages of T- and B-cell development, underscoring that a direct gene correction might capture the precise temporal expression of the endogenous gene. Here, we report a feasibility study using the CRISPR/Cas9-based "universal gene-correction" approach for the RAG2 locus in human hematopoietic stem/progenitor cells (HSPCs) from healthy donors and RAG2-SCID patient. V(D)J-recombinase activity was restored after gene correction of RAG2-SCID-derived HSPCs, resulting in the development of T-cell receptor (TCR) αβ and γδ CD3+ cells and single-positive CD4+ and CD8+ lymphocytes. TCR repertoire analysis indicated a normal distribution of CDR3 length and preserved usage of the distal TRAV genes. We confirmed the in vivo rescue of B-cell development with normal immunoglobulin M surface expression and a significant decrease in CD56bright natural killer cells. Together, we provide specificity, toxicity, and efficacy data supporting the development of a gene-correction therapy to benefit RAG2-deficient patients.
Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution.
Conflict of interest statement
Conflict-of-interest disclosure: M.H.P. holds equity and is on the board of directors of Graphite Bio; is on the scientific advisory board and holds equity in Allogene Therapeutics; and is a founder and holds equity in Kamau Therapeutics. R.M. is on the board of directors of Beyond Spring Inc, and the scientific advisory boards of Zenshine Pharmaceuticals, and Kodikaz Therapeutics Solutions Inc; none of these companies had any input in the design, execution, interpretation, or publication of the work in this manuscript. G.L.K., N.M.B., and C.A.V. are employees of integrated DNA technologies, which offer reagents for sale that are identical or similar to some of the compounds described in the manuscript. The remaining authors declare no competing financial interests.
Figures


Comment in
-
Mending RAG2: gene editing for treatment of RAG2 deficiency.Blood Adv. 2024 Apr 9;8(7):1817-1819. doi: 10.1182/bloodadvances.2023012079. Blood Adv. 2024. PMID: 38592712 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

