Formin tails act as a switch, inhibiting or enhancing processive actin elongation
- PMID: 38097186
- PMCID: PMC10797183
- DOI: 10.1016/j.jbc.2023.105557
Formin tails act as a switch, inhibiting or enhancing processive actin elongation
Abstract
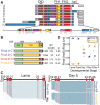
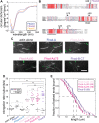
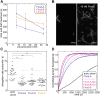
Formins are large, multidomain proteins that nucleate new actin filaments and accelerate elongation through a processive interaction with the barbed ends of filaments. Their actin assembly activity is generally attributed to their eponymous formin homology (FH) 1 and 2 domains; however, evidence is mounting that regions outside of the FH1FH2 stretch also tune actin assembly. Here, we explore the underlying contributions of the tail domain, which spans the sequence between the FH2 domain and the C terminus of formins. Tails vary in length from ∼0 to >200 residues and contain a number of recognizable motifs. The most common and well-studied motif is the ∼15-residue-long diaphanous autoregulatory domain. This domain mediates all or nothing regulation of actin assembly through an intramolecular interaction with the diaphanous inhibitory domain in the N-terminal half of the protein. Multiple reports demonstrate that the tail can enhance both nucleation and processivity. In this study, we provide a high-resolution view of the alternative splicing encompassing the tail in the formin homology domain (Fhod) family of formins during development. While four distinct tails are predicted, we found significant levels of only two of these. We characterized the biochemical effects of the different tails. Surprisingly, the two highly expressed Fhod-tails inhibit processive elongation and diminish nucleation, while a third supports activity. These findings demonstrate a new mechanism of modulating actin assembly by formins and support a model in which splice variants are specialized to build distinct actin structures during development.
Keywords: Drosophila; Fhod; actin; cytoskeleton; formin; long-read sequencing; mRNA.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Goode B.L., Eck M.J. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 2007;76:593–627. - PubMed
-
- Valencia D.A., Quinlan M.E. Formins. Curr. Biol. 2021;31:R517–R522. - PubMed
-
- Xu Y., Moseley J.B., Sagot I., Poy F., Pellman D., Goode B.L., et al. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

