Activation of ILC2s through constitutive IFNγ signaling reduction leads to spontaneous pulmonary fibrosis
- PMID: 38097562
- PMCID: PMC10721793
- DOI: 10.1038/s41467-023-43336-6
Activation of ILC2s through constitutive IFNγ signaling reduction leads to spontaneous pulmonary fibrosis
Abstract
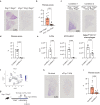
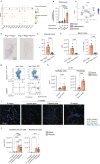
Pulmonary fibrosis (PF), a condition characterized by inflammation and collagen deposition in the alveolar interstitium, causes dyspnea and fatal outcomes. Although the bleomycin-induced PF mouse model has improved our understanding of exogenous factor-induced fibrosis, the mechanism governing endogenous factor-induced fibrosis remains unknown. Here, we find that Ifngr1-/-Rag2-/- mice, which lack the critical suppression factor for group 2 innate lymphoid cells (ILC2), develop PF spontaneously. The onset phase of fibrosis includes ILC2 subpopulations with a high Il1rl1 (IL-33 receptor) expression, and fibrosis does not develop in ILC-deficient or IL-33-deficient mice. Although ILC2s are normally localized near bronchioles and blood vessels, ILC2s are increased in fibrotic areas along with IL-33 positive fibroblasts during fibrosis. Co-culture analysis shows that activated-ILC2s directly induce collagen production from fibroblasts. Furthermore, increased IL1RL1 and decreased IFNGR1 expressions are confirmed in ILC2s from individuals with idiopathic PF, highlighting the applicability of Ifngr1-/-Rag2-/- mice as a mouse model for fibrosis research.
© 2023. The Author(s).
Conflict of interest statement
K. Moro received a grant from Astellas Pharma, Inc. All other authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

