Atherosclerotic plaque development in mice is enhanced by myeloid ZEB1 downregulation
- PMID: 38097578
- PMCID: PMC10721632
- DOI: 10.1038/s41467-023-43896-7
Atherosclerotic plaque development in mice is enhanced by myeloid ZEB1 downregulation
Abstract
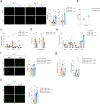
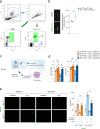
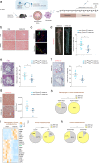
Accumulation of lipid-laden macrophages within the arterial neointima is a critical step in atherosclerotic plaque formation. Here, we show that reduced levels of the cellular plasticity factor ZEB1 in macrophages increase atherosclerotic plaque formation and the chance of cardiovascular events. Compared to control counterparts (Zeb1WT/ApoeKO), male mice with Zeb1 ablation in their myeloid cells (Zeb1∆M/ApoeKO) have larger atherosclerotic plaques and higher lipid accumulation in their macrophages due to delayed lipid traffic and deficient cholesterol efflux. Zeb1∆M/ApoeKO mice display more pronounced systemic metabolic alterations than Zeb1WT/ApoeKO mice, with higher serum levels of low-density lipoproteins and inflammatory cytokines and larger ectopic fat deposits. Higher lipid accumulation in Zeb1∆M macrophages is reverted by the exogenous expression of Zeb1 through macrophage-targeted nanoparticles. In vivo administration of these nanoparticles reduces atherosclerotic plaque formation in Zeb1∆M/ApoeKO mice. Finally, low ZEB1 expression in human endarterectomies is associated with plaque rupture and cardiovascular events. These results set ZEB1 in macrophages as a potential target in the treatment of atherosclerosis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- 2017-SGR-1174 and 2021-SGR-01328/Government of Catalonia | Agència de Gestió d'Ajuts Universitaris i de Recerca (Agency for Management of University and Research Grants)
- PID2020-116338RB-I00/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
- RYC2018-0Z23971-I/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
- 202208110041/China Scholarship Council (CSC)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

