Genome-wide CRISPR screening identifies a role for ARRDC3 in TRP53-mediated responses
- PMID: 38097622
- PMCID: PMC10850147
- DOI: 10.1038/s41418-023-01249-3
Genome-wide CRISPR screening identifies a role for ARRDC3 in TRP53-mediated responses
Abstract
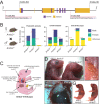
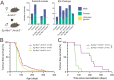
Whole-genome screens using CRISPR technologies are powerful tools to identify novel tumour suppressors as well as factors that impact responses of malignant cells to anti-cancer agents. Applying this methodology to lymphoma cells, we conducted a genome-wide screen to identify novel inhibitors of tumour expansion that are induced by the tumour suppressor TRP53. We discovered that the absence of Arrestin domain containing 3 (ARRDC3) increases the survival and long-term competitiveness of MYC-driven lymphoma cells when treated with anti-cancer agents that activate TRP53. Deleting Arrdc3 in mice caused perinatal lethality due to various developmental abnormalities, including cardiac defects. Notably, the absence of ARRDC3 markedly accelerated MYC-driven lymphoma development. Thus, ARRDC3 is a new mediator of TRP53-mediated suppression of tumour expansion, and this discovery may open new avenues to harness this process for cancer therapy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Lossi L. The concept of intrinsic versus extrinsic apoptosis. Biochem J. 2022;479:357–84. - PubMed
-
- Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–94. - PubMed
-
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–8. - PubMed
-
- Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

