Evolution of lysine-specific demethylase 1 and REST corepressor gene families and their molecular interaction
- PMID: 38097664
- PMCID: PMC10721905
- DOI: 10.1038/s42003-023-05652-x
Evolution of lysine-specific demethylase 1 and REST corepressor gene families and their molecular interaction
Abstract
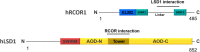
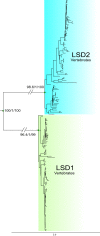
Lysine-specific demethylase 1A (LSD1) binds to the REST corepressor (RCOR) protein family of corepressors to erase transcriptionally active marks on histones. Functional diversity in these complexes depends on the type of RCOR included, which modulates the catalytic activity of the complex. Here, we studied the duplicative history of the RCOR and LSD gene families and analyzed the evolution of their interaction. We found that RCOR genes are the product of the two rounds of whole-genome duplications that occurred early in vertebrate evolution. In contrast, the origin of the LSD genes traces back before to the divergence of animals and plants. Using bioinformatics tools, we show that the RCOR and LSD1 interaction precedes the RCOR repertoire expansion that occurred in the last common ancestor of jawed vertebrates. Overall, we trace LSD1-RCOR complex evolution and propose that animal non-model species offer advantages in addressing questions about the molecular biology of this epigenetic complex.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

