lncRNA-Gm5532 regulates osteoclast differentiation through the miR-125a-3p/TRAF6 axis
- PMID: 38098360
- PMCID: PMC10875346
- DOI: 10.3724/abbs.2023245
lncRNA-Gm5532 regulates osteoclast differentiation through the miR-125a-3p/TRAF6 axis
Abstract
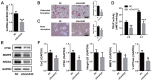
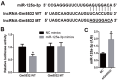
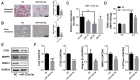
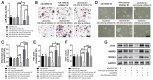
Long noncoding RNAs (lncRNAs) are important regulators of bone metabolism. In this study, lncRNA microarray analysis was used to identify differentially expressed lncRNAs in differentiated osteoclasts. lncRNA-Gm5532 is highly expressed during osteoclast differentiation. lncRNA-Gm5532 knockdown impairs osteoclast formation and bone resorption. Mechanistic experiments show that lncRNA-Gm5532 functions as a competing endogenous RNA (ceRNA) and acts as a sponge for miR-125a-3p, which promotes TNF receptor-associated factor 6 (TRAF6) expression. miR-125a-3p mimics suppress osteoclast differentiation and TAK1/NF-κB/MAPK signaling. The miR-125a-3p inhibitor reverses the negative effects of siGm5532 on osteoclast differentiation. In summary, our study reveals that lncRNA-Gm5532 functions as an activator in osteoclast differentiation by targeting the miR-125a-3p/TRAF6 axis, making it a novel biomarker and potential therapeutic target for osteoporosis.
Keywords: TNF receptor-associated factor 6 (TRAF6); lncRNA-Gm5532; miR-125a-3p; osteoclast differentiation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Extracellular vesicles from mouse bone marrow macrophages-derived osteoclasts treated with zoledronic acid contain miR-146a-5p and miR-322-3p, which inhibit osteoclast function.Bone. 2025 Jan;190:117323. doi: 10.1016/j.bone.2024.117323. Epub 2024 Nov 5. Bone. 2025. PMID: 39510435
-
LncRNA H19 promotes osteoclast differentiation by sponging miR-29c-3p to increase expression of cathepsin K.Bone. 2025 Mar;192:117340. doi: 10.1016/j.bone.2024.117340. Epub 2024 Nov 29. Bone. 2025. PMID: 39615642
-
LncRNA XIST regulates osteoclast formation and promotes orthodontically induced inflammatory root resorption through miR-130b-3p/PTEN axis.Biotechnol Genet Eng Rev. 2024 Nov;40(3):2560-2576. doi: 10.1080/02648725.2023.2200331. Epub 2023 Apr 14. Biotechnol Genet Eng Rev. 2024. PMID: 37057740
-
Tumor-derived exosomal lncRNA-MIR193BHG promotes bone metastasis of breast cancer by targeting the miR-489-3p/DNMT3A signaling axis in osteoclasts.J Transl Med. 2025 Jan 31;23(1):142. doi: 10.1186/s12967-025-06156-4. J Transl Med. 2025. PMID: 39891171 Free PMC article.
-
Advances of long non-coding RNAs in osteoclast differentiation and osteoporosis.Pathol Res Pract. 2024 Aug;260:155413. doi: 10.1016/j.prp.2024.155413. Epub 2024 Jul 6. Pathol Res Pract. 2024. PMID: 38981344 Review.
Cited by
-
TNF receptor-associated factors: promising targets of natural products for the treatment of osteoporosis.Front Physiol. 2025 May 27;16:1527814. doi: 10.3389/fphys.2025.1527814. eCollection 2025. Front Physiol. 2025. PMID: 40496246 Free PMC article. Review.
-
Role of microRNAs in Osteosarcopenic Obesity/Adiposity: A Scoping Review.Cells. 2025 May 29;14(11):802. doi: 10.3390/cells14110802. Cells. 2025. PMID: 40497978 Free PMC article.
References
-
- Reid IR. A broader strategy for osteoporosis interventions. Nat Rev Endocrinol. . 2020;16:333–339. doi: 10.1038/s41574-020-0339-7. - DOI - PubMed
-
- Katsimbri P. The biology of normal bone remodelling. Eur J Cancer Care. . 2017;26:e12740. doi: 10.1111/ecc.12740. - DOI - PubMed
-
- Park-Min KH. Metabolic reprogramming in osteoclasts. Semin Immunopathol. . 2019;41:565–572. doi: 10.1007/s00281-019-00757-0. - DOI - PMC - PubMed
-
- Aurilia C, Donati S, Palmini G, Miglietta F, Iantomasi T, Brandi ML. The involvement of long non-coding RNAs in bone. Int J Mol Sci. . 2021;22:3909. doi: 10.3390/ijms22083909. - DOI - PMC - PubMed
-
- Liu W, Li Z, Cai Z, Xie Z, Li J, Li M, Cen S, et al. LncRNA‐mRNA expression profiles and functional networks in osteoclast differentiation. J Cell Mol Medi. . 2020;24:9786–9797. doi: 10.1111/jcmm.15560. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

