Combining the Tyrosine Kinase Inhibitor Cabozantinib and the mTORC1/2 Inhibitor Sapanisertib Blocks ERK Pathway Activity and Suppresses Tumor Growth in Renal Cell Carcinoma
- PMID: 38098449
- PMCID: PMC10722140
- DOI: 10.1158/0008-5472.CAN-23-0604
Combining the Tyrosine Kinase Inhibitor Cabozantinib and the mTORC1/2 Inhibitor Sapanisertib Blocks ERK Pathway Activity and Suppresses Tumor Growth in Renal Cell Carcinoma
Abstract
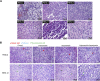
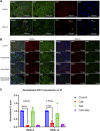
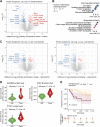
Current treatment approaches for renal cell carcinoma (RCC) face challenges in achieving durable tumor responses due to tumor heterogeneity and drug resistance. Combination therapies that leverage tumor molecular profiles could offer an avenue for enhancing treatment efficacy and addressing the limitations of current therapies. To identify effective strategies for treating RCC, we selected ten drugs guided by tumor biology to test in six RCC patient-derived xenograft (PDX) models. The multitargeted tyrosine kinase inhibitor (TKI) cabozantinib and mTORC1/2 inhibitor sapanisertib emerged as the most effective drugs, particularly when combined. The combination demonstrated favorable tolerability and inhibited tumor growth or induced tumor regression in all models, including two from patients who experienced treatment failure with FDA-approved TKI and immunotherapy combinations. In cabozantinib-treated samples, imaging analysis revealed a significant reduction in vascular density, and single-nucleus RNA sequencing (snRNA-seq) analysis indicated a decreased proportion of endothelial cells in the tumors. SnRNA-seq data further identified a tumor subpopulation enriched with cell-cycle activity that exhibited heightened sensitivity to the cabozantinib and sapanisertib combination. Conversely, activation of the epithelial-mesenchymal transition pathway, detected at the protein level, was associated with drug resistance in residual tumors following combination treatment. The combination effectively restrained ERK phosphorylation and reduced expression of ERK downstream transcription factors and their target genes implicated in cell-cycle control and apoptosis. This study highlights the potential of the cabozantinib plus sapanisertib combination as a promising treatment approach for patients with RCC, particularly those whose tumors progressed on immune checkpoint inhibitors and other TKIs.
Significance: The molecular-guided therapeutic strategy of combining cabozantinib and sapanisertib restrains ERK activity to effectively suppress growth of renal cell carcinomas, including those unresponsive to immune checkpoint inhibitors.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures





References
-
- Bedke J, Albiges L, Capitanio U, Giles RH, Hora M, Lam TB, et al. . The 2021 updated European association of urology guidelines on renal cell carcinoma: immune checkpoint inhibitor–based combination therapies for treatment-naive metastatic clear-cell renal cell carcinoma are standard of care. Eur Urol 2021;80:393–7. - PubMed
-
- Motzer R, Alekseev B, Rha S-Y, Porta C, Eto M, Powles T, et al. . Lenvatinib plus pembrolizumab or everolimus for advanced renal cell carcinoma. N Engl J Med 2021;384:1289–300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

