Advances in transposable elements: from mechanisms to applications in mammalian genomics
- PMID: 38098473
- PMCID: PMC10719622
- DOI: 10.3389/fgene.2023.1290146
Advances in transposable elements: from mechanisms to applications in mammalian genomics
Abstract
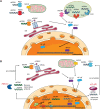
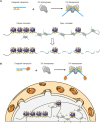
It has been 70 years since Barbara McClintock discovered transposable elements (TE), and the mechanistic studies and functional applications of transposable elements have been at the forefront of life science research. As an essential part of the genome, TEs have been discovered in most species of prokaryotes and eukaryotes, and the relative proportion of the total genetic sequence they comprise gradually increases with the expansion of the genome. In humans, TEs account for about 40% of the genome and are deeply involved in gene regulation, chromosome structure maintenance, inflammatory response, and the etiology of genetic and non-genetic diseases. In-depth functional studies of TEs in mammalian cells and the human body have led to a greater understanding of these fundamental biological processes. At the same time, as a potent mutagen and efficient genome editing tool, TEs have been transformed into biological tools critical for developing new techniques. By controlling the random insertion of TEs into the genome to change the phenotype in cells and model organisms, critical proteins of many diseases have been systematically identified. Exploiting the TE's highly efficient in vitro insertion activity has driven the development of cutting-edge sequencing technologies. Recently, a new technology combining CRISPR with TEs was reported, which provides a novel targeted insertion system to both academia and industry. We suggest that interrogating biological processes that generally depend on the actions of TEs with TEs-derived genetic tools is a very efficient strategy. For example, excessive activation of TEs is an essential factor in the occurrence of cancer in humans. As potent mutagens, TEs have also been used to unravel the key regulatory elements and mechanisms of carcinogenesis. Through this review, we aim to effectively combine the traditional views of TEs with recent research progress, systematically link the mechanistic discoveries of TEs with the technological developments of TE-based tools, and provide a comprehensive approach and understanding for researchers in different fields.
Keywords: RNA-guided transposition; aging; forward genetic screening; high throughput sequencing; immune response; transposable elements; tumor genetics.
Copyright © 2023 Han, Perkins, Novaes, Xu and Chang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

