Regulatory mechanisms triggered by enzyme interactions with lipid membrane surfaces
- PMID: 38099197
- PMCID: PMC10720463
- DOI: 10.3389/fmolb.2023.1306483
Regulatory mechanisms triggered by enzyme interactions with lipid membrane surfaces
Abstract
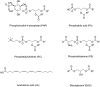
Recruitment of enzymes to intracellular membranes often modulates their catalytic activity, which can be important in cell signaling and membrane trafficking. Thus, re-localization is not only important for these enzymes to gain access to their substrates, but membrane interactions often allosterically regulate enzyme function by inducing conformational changes across different time and amplitude scales. Recent structural, biophysical and computational studies have revealed how key enzymes interact with lipid membrane surfaces, and how this membrane binding regulates protein structure and function. This review summarizes the recent progress in understanding regulatory mechanisms involved in enzyme-membrane interactions.
Keywords: allosteric regulation; conformational change; lipid kinase; lipid metabolism; phosphatidylinositol phosphate lipids; protein-membrane interactions; signaling.
Copyright © 2023 Yu and Boehr.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

