Foxp3-mediated blockage of ryanodine receptor 2 underlies contact-based suppression by regulatory T cells
- PMID: 38099494
- PMCID: PMC10721146
- DOI: 10.1172/JCI163470
Foxp3-mediated blockage of ryanodine receptor 2 underlies contact-based suppression by regulatory T cells
Abstract
The suppression mechanism of Tregs remains an intensely investigated topic. As our focus has shifted toward a model centered on indirect inhibition of DCs, a universally applicable effector mechanism controlled by the transcription factor forkhead box P3 (Foxp3) expression has not been found. Here, we report that Foxp3 blocked the transcription of ER Ca2+-release channel ryanodine receptor 2 (RyR2). Reduced RyR2 shut down basal Ca2+ oscillation in Tregs, which reduced m-calpain activities that are needed for T cells to disengage from DCs, suggesting a persistent blockage of DC antigen presentation. RyR2 deficiency rendered the CD4+ T cell pool immune suppressive and caused it to behave in the same manner as Foxp3+ Tregs in viral infection, asthma, hypersensitivity, colitis, and tumor development. In the absence of Foxp3, Ryr2-deficient CD4+ T cells rescued the systemic autoimmunity associated with scurfy mice. Therefore, Foxp3-mediated Ca2+ signaling inhibition may be a central effector mechanism of Treg immune suppression.
Keywords: Antigen-presenting cells; Autoimmunity; Immunology; Tolerance.
Figures


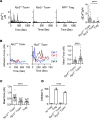
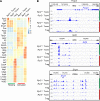
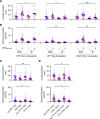

Comment in
-
Strengthening bonds via RyR2 inhibition helps immune suppression.J Clin Invest. 2023 Dec 15;133(24):e172986. doi: 10.1172/JCI172986. J Clin Invest. 2023. PMID: 38099491 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

