Genome sequence-based identification of Enterobacter strains and description of Enterobacter pasteurii sp. nov
- PMID: 38099614
- PMCID: PMC10783019
- DOI: 10.1128/spectrum.03150-23
Genome sequence-based identification of Enterobacter strains and description of Enterobacter pasteurii sp. nov
Abstract
Accurate taxonomy is essential for microbial biological resource centers, since the microbial resources are often used to support new discoveries and subsequent research. Here, we used genome sequence data, alongside matrix-assisted laser desorption/ionization time-of-flight mass spectrometer biotyper-based protein profiling, to accurately identify six Enterobacter cloacae complex strains. This approach effectively identified distinct species within the E. cloacae complex, including Enterobacter asburiae, "Enterobacter xiangfangensis," and Enterobacter quasihormaechei. Moreover, the study revealed the existence of a novel species within the Enterobacter genus, for which we proposed the name Enterobacter pasteurii sp. nov. In summary, this study demonstrates the significance of adopting a genome sequence-driven taxonomy approach for the precise identification of bacterial strains in a biological resource center and expands our understanding of the E. cloacae complex.
Keywords: Enterobacter; Enterobacteriaceae; MALDI-TOF MS; phylogenomic; taxonomy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
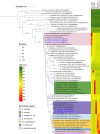

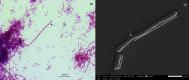

References
-
- Dong X, Zhu M, Li Y, Huang D, Wang L, Yan C, Zhang L, Dong F, Lu J, Lin X, Li K, Bao Q, Cong C, Pan W. 2022. Whole-genome sequencing-based species classification, multilocus sequence typing, and antimicrobial resistance mechanism analysis of the Enterobacter Cloacae complex in Southern China. Microbiol Spectr 10:e0216022. doi:10.1128/spectrum.02160-22 - DOI - PMC - PubMed
-
- World Health Organization . 2018. Global antimicrobial resistance surveillance system (GLASS) report: early implementation 2017-2018. World Health Organization, Geneva. https://iris.who.int/handle/10665/279656.
-
- Hormaeche E, Edwards PR. 1960. A proposed genus Enterobacter. Internat Bull Bacteriol Nomencla Taxon 10:71–74. doi:10.1099/0096266X-10-2-71 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

