Chemistry and biology of specialized metabolites produced by Actinomadura
- PMID: 38099919
- PMCID: PMC10951976
- DOI: 10.1039/d3np00047h
Chemistry and biology of specialized metabolites produced by Actinomadura
Abstract
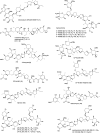
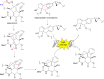
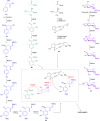
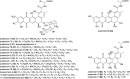
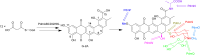
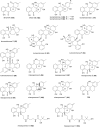
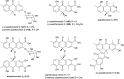
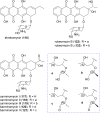
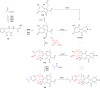
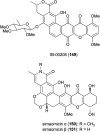
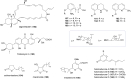
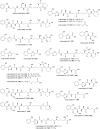
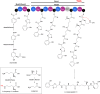
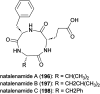
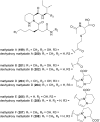
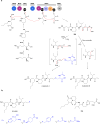
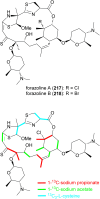
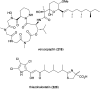
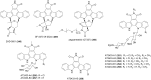
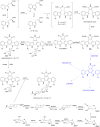
Covering: up to the end of 2022In recent years rare Actinobacteria have become increasingly recognised as a rich source of novel bioactive metabolites. Actinomadura are Gram-positive bacteria that occupy a wide range of ecological niches. This review highlights about 230 secondary metabolites produced by Actinomadura spp., reported until the end of 2022, including their bioactivities and selected biosynthetic pathways. Notably, the bioactive compounds produced by Actinomadura spp. demonstrate a wide range of activities, including antimicrobial, antitumor and anticoccidial effects, highlighting their potential in various fields.
Conflict of interest statement
There is no conflicts to declare.
Figures











References
-
- Michael Goodfellow P. K., Busse H.-J., Trujillo M. E., Suzuki K.-I., Ludwig W. and Whitman W. B., Bergey's Manual of Systematic Bacteriology: Volume 5: The Actinobacteria, Springer, New York, NY, 2 edn, 2019
-
- Hopwood D. A., Streptomyces in Nature and Medicine: The Antibiotic Makers, Oxford University Press, 2007
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

