Kinetic evidence for multiple aggregation pathways in antibody light chain variable domains
- PMID: 38100259
- PMCID: PMC10868443
- DOI: 10.1002/pro.4871
Kinetic evidence for multiple aggregation pathways in antibody light chain variable domains
Abstract
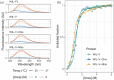
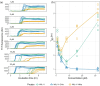
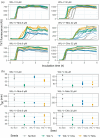
Aggregation of antibody light chain proteins is associated with the progressive disease light chain amyloidosis. Patient-derived amyloid fibrils are formed from light chain variable domain residues in non-native conformations, highlighting a requirement that light chains unfold from their native structures in order to aggregate. However, mechanistic studies of amyloid formation have primarily focused on the self-assembly of natively unstructured peptides, and the role of native state unfolding is less well understood. Using a well-studied light chain variable domain protein known as WIL, which readily aggregates in vitro under conditions where the native state predominates, we asked how the protein concentration and addition of pre-formed fibril "seeds" alter the kinetics of aggregation. Monitoring aggregation with thioflavin T fluorescence revealed a distinctly non-linear dependence on concentration, with a maximum aggregation rate observed at 8 μM protein. This behavior is consistent with formation of alternate aggregate structures in the early phases of amyloid formation. Addition of N- or C-terminal peptide tags, which did not greatly affect the folding or stability of the protein, altered the concentration dependence of aggregation. Aggregation rates increased in the presence of pre-formed seeds, but this effect did not eliminate the delay before aggregation and became saturated when the proportion of seeds added was greater than 1 in 1600. The complexity of aggregation observed in vitro highlights how multiple species may contribute to amyloid pathology in patients.
Keywords: AL amyloidosis; aggregation kinetics; amyloid fibrils; antibody variable domain; protein misfolding; systemic light chain amyloidosis.
© 2023 The Protein Society.
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
Kinetic evidence for multiple aggregation pathways in antibody light chain variable domains.bioRxiv [Preprint]. 2023 Aug 28:2023.08.28.555139. doi: 10.1101/2023.08.28.555139. bioRxiv. 2023. Update in: Protein Sci. 2024 Mar;33(3):e4871. doi: 10.1002/pro.4871. PMID: 37693524 Free PMC article. Updated. Preprint.
Similar articles
-
Kinetic evidence for multiple aggregation pathways in antibody light chain variable domains.bioRxiv [Preprint]. 2023 Aug 28:2023.08.28.555139. doi: 10.1101/2023.08.28.555139. bioRxiv. 2023. Update in: Protein Sci. 2024 Mar;33(3):e4871. doi: 10.1002/pro.4871. PMID: 37693524 Free PMC article. Updated. Preprint.
-
The Kinetic Stability of a Full-Length Antibody Light Chain Dimer Determines whether Endoproteolysis Can Release Amyloidogenic Variable Domains.J Mol Biol. 2016 Oct 23;428(21):4280-4297. doi: 10.1016/j.jmb.2016.08.021. Epub 2016 Aug 26. J Mol Biol. 2016. PMID: 27569045 Free PMC article.
-
Partially folded intermediates as critical precursors of light chain amyloid fibrils and amorphous aggregates.Biochemistry. 2001 Mar 27;40(12):3525-35. doi: 10.1021/bi001782b. Biochemistry. 2001. PMID: 11297418
-
Immunoglobulin light chain amyloid aggregation.Chem Commun (Camb). 2018 Sep 20;54(76):10664-10674. doi: 10.1039/c8cc04396e. Chem Commun (Camb). 2018. PMID: 30087961 Free PMC article. Review.
-
Dynamic protein structures in normal function and pathologic misfolding in systemic amyloidosis.Biophys Chem. 2022 Jan;280:106699. doi: 10.1016/j.bpc.2021.106699. Epub 2021 Oct 14. Biophys Chem. 2022. PMID: 34773861 Free PMC article. Review.
References
-
- Buxbaum JN, Dispenzieri A, Eisenberg DS, Fändrich M, Merlini G, Saraiva MJM, et al. Amyloid nomenclature 2022: update, novel proteins, and recommendations by the International Society of Amyloidosis (ISA) Nomenclature Committee. Amyloid. 2022;29:213–219. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

