FRAGSITE2: A structure and fragment-based approach for virtual ligand screening
- PMID: 38100293
- PMCID: PMC10751727
- DOI: 10.1002/pro.4869
FRAGSITE2: A structure and fragment-based approach for virtual ligand screening
Abstract
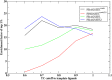
Protein function annotation and drug discovery often involve finding small molecule binders. In the early stages of drug discovery, virtual ligand screening (VLS) is frequently applied to identify possible hits before experimental testing. While our recent ligand homology modeling (LHM)-machine learning VLS method FRAGSITE outperformed approaches that combined traditional docking to generate protein-ligand poses and deep learning scoring functions to rank ligands, a more robust approach that could identify a more diverse set of binding ligands is needed. Here, we describe FRAGSITE2 that shows significant improvement on protein targets lacking known small molecule binders and no confident LHM identified template ligands when benchmarked on two commonly used VLS datasets: For both the DUD-E set and DEKOIS2.0 set and ligands having a Tanimoto coefficient (TC) < 0.7 to the template ligands, the 1% enrichment factor (EF1% ) of FRAGSITE2 is significantly better than those for FINDSITEcomb2.0 , an earlier LHM algorithm. For the DUD-E set, FRAGSITE2 also shows better ROC enrichment factor and AUPR (area under the precision-recall curve) than the deep learning DenseFS scoring function. Comparison with the RF-score-VS on the 76 target subset of DEKOIS2.0 and a TC < 0.99 to training DUD-E ligands, FRAGSITE2 has double the EF1% . Its boosted tree regression method provides for more robust performance than a deep learning multiple layer perceptron method. When compared with the pretrained language model for protein target features, FRAGSITE2 also shows much better performance. Thus, FRAGSITE2 is a promising approach that can discover novel hits for protein targets. FRAGSITE2's web service is freely available to academic users at http://sites.gatech.edu/cssb/FRAGSITE2.
Keywords: FINDSITE; FRAGSITE; drug discovery; ligand homology modeling; machine learning; template ligand; virtual ligand screening.
© 2023 The Protein Society.
Figures


References
-
- Anonymous . Daylight theory manual. Aliso Viejo, CA: Daylight Chemical Information Systems, Inc; 2007.
-
- Bauer MR, Ibrahim TM, Vogel SM, Boeckler FM. Evaluation and optimization of virtual screening workflows with DEKOIS 2.0 – a public library of challenging docking benchmark sets. J Chem Inf Model. 2013;53:1447–1462. - PubMed
-
- Bernstein FC, Koetzle TF, Williams GJB, Meyer EF Jr, Brice MD, Rodgers JR, et al. The Protein Data Bank: a computer‐based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

