A study of deep active learning methods to reduce labelling efforts in biomedical relation extraction
- PMID: 38100453
- PMCID: PMC10723703
- DOI: 10.1371/journal.pone.0292356
A study of deep active learning methods to reduce labelling efforts in biomedical relation extraction
Abstract
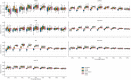
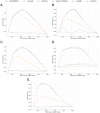
Automatic biomedical relation extraction (bioRE) is an essential task in biomedical research in order to generate high-quality labelled data that can be used for the development of innovative predictive methods. However, building such fully labelled, high quality bioRE data sets of adequate size for the training of state-of-the-art relation extraction models is hindered by an annotation bottleneck due to limitations on time and expertise of researchers and curators. We show here how Active Learning (AL) plays an important role in resolving this issue and positively improve bioRE tasks, effectively overcoming the labelling limits inherent to a data set. Six different AL strategies are benchmarked on seven bioRE data sets, using PubMedBERT as the base model, evaluating their area under the learning curve (AULC) as well as intermediate results measurements. The results demonstrate that uncertainty-based strategies, such as Least-Confident or Margin Sampling, are statistically performing better in terms of F1-score, accuracy and precision, than other types of AL strategies. However, in terms of recall, a diversity-based strategy, called Core-set, outperforms all strategies. AL strategies are shown to reduce the annotation need (in order to reach a performance at par with training on all data), from 6% to 38%, depending on the data set; with Margin Sampling and Least-Confident Sampling strategies moreover obtaining the best AULCs compared to the Random Sampling baseline. We show through the experiments the importance of using AL methods to reduce the amount of labelling needed to construct high-quality data sets leading to optimal performance of deep learning models. The code and data sets to reproduce all the results presented in the article are available at https://github.com/oligogenic/Deep_active_learning_bioRE.
Copyright: © 2023 Nachtegael et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Liu H, Friedman C. Mining terminological knowledge in large biomedical corpora. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing. 2003; p. 415–426. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

