Acquisition of suppressive function by conventional T cells limits antitumor immunity upon Treg depletion
- PMID: 38100544
- PMCID: PMC7615475
- DOI: 10.1126/sciimmunol.abo5558
Acquisition of suppressive function by conventional T cells limits antitumor immunity upon Treg depletion
Erratum in
-
Erratum for the Research Article "Acquisition of suppressive function by conventional T cells limits antitumor immunity upon Treg depletion" by S. K. Whiteside et al.Sci Immunol. 2024 Dec 13;9(102):eadu6398. doi: 10.1126/sciimmunol.adu6398. Epub 2024 Dec 13. Sci Immunol. 2024. PMID: 39671471 No abstract available.
Abstract
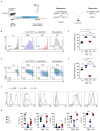
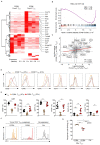
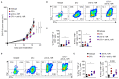
Regulatory T (Treg) cells contribute to immune homeostasis but suppress immune responses to cancer. Strategies to disrupt Treg cell-mediated cancer immunosuppression have been met with limited clinical success, but the underlying mechanisms for treatment failure are poorly understood. By modeling Treg cell-targeted immunotherapy in mice, we find that CD4+ Foxp3- conventional T (Tconv) cells acquire suppressive function upon depletion of Foxp3+ Treg cells, limiting therapeutic efficacy. Foxp3- Tconv cells within tumors adopt a Treg cell-like transcriptional profile upon ablation of Treg cells and acquire the ability to suppress T cell activation and proliferation ex vivo. Suppressive activity is enriched among CD4+ Tconv cells marked by expression of C-C motif receptor 8 (CCR8), which are found in mouse and human tumors. Upon Treg cell depletion, CCR8+ Tconv cells undergo systemic and intratumoral activation and expansion, and mediate IL-10-dependent suppression of antitumor immunity. Consequently, conditional deletion of Il10 within T cells augments antitumor immunity upon Treg cell depletion in mice, and antibody blockade of IL-10 signaling synergizes with Treg cell depletion to overcome treatment resistance. These findings reveal a secondary layer of immunosuppression by Tconv cells released upon therapeutic Treg cell depletion and suggest that broader consideration of suppressive function within the T cell lineage is required for development of effective Treg cell-targeted therapies.
Conflict of interest statement
R.R. holds or has held paid consultancies with Lyell Immunopharma, Achilles Therapeutics and Enhanc3D Genomics; and is a principal investigator of research projects funded by AstraZeneca and F-star Therapeutics on unrelated topics that do not constitute competing interests. E.L. served as a consltant for BD Biosciences on a topic unrelated to this work. All other authors declare no competing interests.
Figures




References
-
- Robert C, Thomas L, Bondarenko I, O’Day S, Weber J, Garbe C, Lebbe C, Baurain JF, Testori A, Grob JJ, Davidson N, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364:2517–2526. - PubMed
-
- Larkin J, Lao CD, Urba WJ, McDermott DF, Horak C, Jiang J, Wolchok JD. Efficacy and Safety of Nivolumab in Patients With BRAF V600 Mutant and BRAF Wild-Type Advanced Melanoma: A Pooled Analysis of 4 Clinical Trials. JAMA Oncol. 2015;1:433–440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

