Single-Cell Profiling Indicates a Proinflammatory Role of Meningeal Ectopic Lymphoid Tissue in Experimental Autoimmune Encephalomyelitis
- PMID: 38100739
- PMCID: PMC10723639
- DOI: 10.1212/NXI.0000000000200185
Single-Cell Profiling Indicates a Proinflammatory Role of Meningeal Ectopic Lymphoid Tissue in Experimental Autoimmune Encephalomyelitis
Abstract
Background and objectives: The factors that drive progression in multiple sclerosis (MS) remain obscure. Identification of key properties of meningeal inflammation will contribute to a better understanding of the mechanisms of progression and how to prevent it.
Methods: Applying single-cell RNA sequencing, we compared gene expression profiles in immune cells from meningeal ectopic lymphoid tissue (mELT) with those from secondary lymphoid organs (SLOs) in spontaneous chronic experimental autoimmune encephalomyelitis (EAE), an animal model of MS.
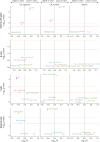
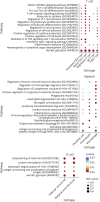
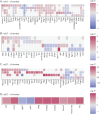
Results: Generally, mELT contained the same immune cell types as SLOs, suggesting a close relationship. Preponderance of B cells over T cells, an increase in regulatory T cells and granulocytes, and a decrease in naïve CD4+ T cells characterize mELT compared with SLOs. Differential gene expression analysis revealed that immune cells in mELT show a more activated and proinflammatory phenotype compared with their counterparts in SLOs. However, the increase in regulatory T cells and upregulation of immunosuppressive genes in most immune cell types indicate that there are mechanisms in place to counter-regulate the inflammatory events, keeping the immune response emanating from mELT in check.
Discussion: Common features in immune cell composition and gene expression indicate that mELT resembles SLOs and may be regarded as a tertiary lymphoid tissue. Distinct differences in expression profiles suggest that mELT rather than SLOs is a key driver of CNS inflammation in spontaneous EAE. Our data provide a starting point for further exploration of molecules or pathways that could be targeted to disrupt mELT formation.
Conflict of interest statement
J. Diddens, G. Lepennetier, V. Friedrich, M. Pfaller, and T. Georgieva report no disclosures relevant to the manuscript. R.M. Brand is a fellow of the Hertie Foundation (medMS Doctoral Program). B. Hemmer has served on scientific advisory boards for Novartis and Sandoz; he has served as DMSC member for AllergyCare, Sandoz, Polpharma, Biocon, and TG therapeutics; his institution received research grants from Roche for multiple sclerosis research. He has received honoraria for counseling (Gerson Lehrmann Group). He holds part of 2 patents; one for the detection of antibodies against KIR4.1 in a subpopulation of patients with multiple sclerosis and the other for genetic determinants of neutralizing antibodies to interferon. All conflicts are not relevant to the topic of the study. K. Lehmann-Horn has received research support (to TUM) from Novartis and honoraria and compensation for travel expenses from Novartis, F. Hoffmann-La Roche, Biogen, Teva, Hexal, and Merck Serono. Go to
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
