Immunoglobulin-like receptors in chickens: identification, functional characterization, and renaming to cluster homolog of immunoglobulin-like receptors
- PMID: 38100950
- PMCID: PMC10764270
- DOI: 10.1016/j.psj.2023.103292
Immunoglobulin-like receptors in chickens: identification, functional characterization, and renaming to cluster homolog of immunoglobulin-like receptors
Abstract
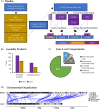
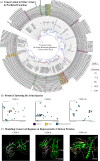
The cluster homolog of immunoglobulin-like receptors (CHIRs), previously known as the "chicken homolog of immunogloublin-like receptors," represents is a large group of transmembrane glycoproteins that direct the immune response. However, the full repertoire of putatively activating, inhibitory, or dual function CHIRA, CHIRB, and CHIRAB on chickens' immune responses is poorly understood. Herein, the study objective was to determine the genes encoding CHIR proteins and predict their function by searching canonical protein structure. A bioinformatics pipeline based on previous work was employed to search for the CHIRs from the newly updated broiler and layer genomes. The categorization into CHIRA, CHIRB, and CHIRAB types was assigned through motif searches, multiple sequence alignment, and phylogeny. In total, 150 protein-encoding genes on Chromosome 31 were identified as CHIRs. Gene members of each functional group (CHIRA, CHIRB, CHIRAB) were classified in accordance with previously recognized proteins. The genes were renamed to "cluster homolog of immunoglobulin-like receptors" (CHIRs) to allow for the naming of orthologous genes in other avian species. Additionally, expression analysis of the classified CHIRs across various reinforces their importance as immune regulators and activation in inflammatory tissues. Furthermore, over 1,000 diverse and rare CHIRs variants associated with differential Marek's disease response (P < 0.05) emphasize the impact of CHIRs on shaping avian immune responses in diverse contexts. The practical applications of these findings encompass advancing immunology, improving poultry health management, optimizing breeding programs for disease resistance, and enhancing overall animal health through a deeper understanding of the roles and functions of CHIRA, CHIRB, and CHIRAB types in avian immune responses.
Keywords: bioinformatics; chicken; cluster homolog of immunoglobulin-like receptors; genome and transcriptome; immunogenetics.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Allaire J. Boston, MA; 2012. RStudio: integrated development environment for R.https://www.posit.co/ Accessed Dec. 2023.
-
- Andrews S. Babraham Bioinformatics, Babraham Institute; Cambridge, UK: 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data.
-
- Bacon L.D., Hunt H.D., Cheng H.H. A review of the development of chicken lines to resolve genes determining resistance to diseases. Poult. Sci. 2000;79:1082–1093. - PubMed
-
- Bacon L.D., Witter R.L. Influence of turkey herpesvirus vaccination on the B-haplotype effect on Marek’s disease resistance in 15. B-congenic chickens. Avian Dis. 1992;36:378–385. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

