Analysis of multiple programmed cell death-related prognostic genes and functional validations of necroptosis-associated genes in oesophageal squamous cell carcinoma
- PMID: 38101299
- PMCID: PMC10733113
- DOI: 10.1016/j.ebiom.2023.104920
Analysis of multiple programmed cell death-related prognostic genes and functional validations of necroptosis-associated genes in oesophageal squamous cell carcinoma
Abstract
Background: Oesophageal squamous cell carcinoma (ESCC) is a lethal malignancy. Immune checkpoint inhibitors (ICIs) showed great clinical benefits for patients with ESCC. We aimed to construct a model predicting prognosis and response to ICIs by integrating diverse programmed cell death (PCD) forms.
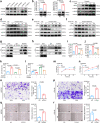
Methods: Genes related to 14 PCDs were collected to generate multi-gene signatures, including apoptosis, necroptosis, pyroptosis, ferroptosis, and cuproptosis. Bulk and single-cell RNA transcriptome datasets were used to develop and validate the model. We assessed the functions of two necroptosis-related genes in ESCC cells by Western blot, co-immunoprecipitation (Co-IP), LDH release assay, CCK-8, and migration assay, followed by immunohistochemistry (IHC) staining on samples of patients with ESCC (n = 67).
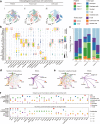
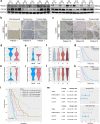
Findings: We built and validated a 16-gene prognostic combined cell death index (CCDI) by combining immunogenic cell death (ICD) and necroptosis signatures. The CCDI could also predict response to ICIs in cancer, as shown by Tumour Immune Dysfunction and Exclusion (TIDE) analysis, confirmed in four independent ICI clinical trials. Trajectory analysis revealed that HOOK1 and CUL4A might affect ESCC cell fate. We found that HOOK1 induced necroptosis and inhibited the proliferation and migration of ESCC cells, while CUL4A exhibited the opposite effects. Co-IP assay confirmed that HOOK1 and CUL4A promoted and reduced necrosome formation in ESCC cells. Data from patients with ESCC further supported that HOOK1 and CUL4A might be a tumour suppressor and oncogene, respectively.
Interpretation: We constructed a CCDI model with potential in predicting prognosis and response to ICIs in cancer. HOOK1 and CUL4A in the CCDI model are crucial prognostic biomarkers in ESCC.
Funding: The Natural Science Foundation of China [82172786], The National Cancer Center Climbing Fund of China [NCC201908B06], The Natural Science Foundation of Heilongjiang Province [LH2021H077].
Keywords: ESCC; Immune checkpoint inhibitor; Programmed cell death; Single-cell RNA-Seq.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests There was no competing interest to disclose.
Figures





References
-
- Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. - PubMed
-
- Overholtzer M., Mailleux A.A., Mouneimne G., et al. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 2007;131(5):966–979. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

