ChREBP is activated by reductive stress and mediates GCKR-associated metabolic traits
- PMID: 38101397
- PMCID: PMC10842884
- DOI: 10.1016/j.cmet.2023.11.010
ChREBP is activated by reductive stress and mediates GCKR-associated metabolic traits
Abstract
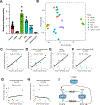
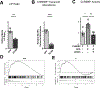
Common genetic variants in glucokinase regulator (GCKR), which encodes GKRP, a regulator of hepatic glucokinase (GCK), influence multiple metabolic traits in genome-wide association studies (GWASs), making GCKR one of the most pleiotropic GWAS loci in the genome. It is unclear why. Prior work has demonstrated that GCKR influences the hepatic cytosolic NADH/NAD+ ratio, also referred to as reductive stress. Here, we demonstrate that reductive stress is sufficient to activate the transcription factor ChREBP and necessary for its activation by the GKRP-GCK interaction, glucose, and ethanol. We show that hepatic reductive stress induces GCKR GWAS traits such as increased hepatic fat, circulating FGF21, and circulating acylglycerol species, which are also influenced by ChREBP. We define the transcriptional signature of hepatic reductive stress and show its upregulation in fatty liver disease and downregulation after bariatric surgery in humans. These findings highlight how a GCKR-reductive stress-ChREBP axis influences multiple human metabolic traits.
Keywords: ChREBP; FGF21; GCK; GCKR; MLIXPL; NAD(+); NADH; fatty liver disease; gastric bypass surgery; metabolism; reductive stress; trigylcerides.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests V.K.M. and V.C. are listed as inventors on a patent application filed by Massachusetts General Hospital on the therapeutic uses of LbNOX. V.K.M. is a scientific advisor to and receives equity from 5AM Ventures. A.C.M. received research support from Boehringer Ingelheim and GlaxoSmithKline for other projects not related to this work.
Figures





References
-
- Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and N.I. of B.R., Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PIW, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, et al. (2007). Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316, 1331–1336. 10.1126/science.1142358. - DOI - PubMed
-
- Beer NL, Tribble ND, McCulloch LJ, Roos C, Johnson PRV, Orho-Melander M, and Gloyn AL (2009). The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet 18, 4081–4088. 10.1093/hmg/ddp357. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

