Ginsenoside Rb2 inhibits p300-mediated SF3A2 acetylation at lysine 10 to promote Fscn1 alternative splicing against myocardial ischemic/reperfusion injury
- PMID: 38101749
- PMCID: PMC11518965
- DOI: 10.1016/j.jare.2023.12.012
Ginsenoside Rb2 inhibits p300-mediated SF3A2 acetylation at lysine 10 to promote Fscn1 alternative splicing against myocardial ischemic/reperfusion injury
Abstract
Introduction: Ginsenosides (GS) derived from Panax ginseng can regulate protein acetylation to promote mitochondrial function for protecting cardiomyocytes. However, the potential mechanisms of GS for regulating acetylation modification are not yet clear.
Objectives: This study aimed to explore the potential mechanisms of GS in regulating protein acetylation and identify ginsenoside monomer for fighting myocardial ischemia-related diseases.
Methods: The 4D-lable free acetylomic analysis was employed to gain the acetylated proteins regulated by GS pretreatment. The co-immunoprecipitation assay, immunofluorescent staining, and mitochondrial respiration measurement were performed to detect the effect of GS or ginsenoside monomer on acetylated protein level and mitochondrial function. RNA sequencing, site-specific mutation, and shRNA interference were used to explore the downstream targets of acetylation modificationby GS. Cellular thermal shift assay and surface plasmon resonance were used for identifying the binding of ginsenoside with target protein.
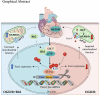
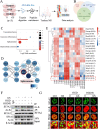
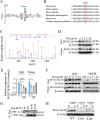
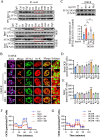
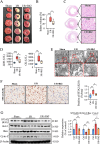
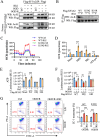
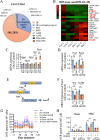
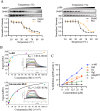
Results: In the cardiomyocytes of normal, oxygen glucose deprivation and/or reperfusion conditions, the acetylomic analysis identified that the acetylated levels of spliceosome proteins were inhibited by GS pretreatment and SF3A2 acetylation at lysine 10 (K10) was significantly decreased as a potential target of GS. Ginsenoside Rb2 was identified as one of the active ginsenoside monomers for reducing the acetylation of SF3A2 (K10), which enhanced mitochondrial respiration against myocardial ischemic injury in in vivo and in vitro experiments. RNA-seq analysis showed that ginsenoside Rb2 promoted alternative splicing of mitochondrial function-related genes and the level of fascin actin-bundling protein 1 (Fscn1) was obviously upregulated, which was dependent on SF3A2 acetylation. Critically, thermodynamic, kinetic and enzymatic experiments demonstrated that ginsenoside Rb2 directly interacted with p300 for inhibiting its activity.
Conclusion: These findings provide a novel mechanism underlying cardiomyocyte protection of ginsenoside Rb2 by inhibiting p300-mediated SF3A2 acteylation for promoting Fscn1 expression, which might be a promising approach for the prevention and treatment of myocardial ischemic diseases.
Keywords: Acetylation; Ginsenoside; Mitochondrial function; Myocardial ischemic/reperfusion injury; P300; SF3A2.
Copyright © 2024. Production and hosting by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Zhang Z., Dalan R., Hu Z., Wang J., Chew N., Poh K., et al. Reactive oxygen species scavenging nanomedicine for the treatment of ischemic heart disease. Adv Mater (Deerfield Beach, Fla) 2022;34:e2202169. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

