Rho GTPase activity crosstalk mediated by Arhgef11 and Arhgef12 coordinates cell protrusion-retraction cycles
- PMID: 38102112
- PMCID: PMC10724141
- DOI: 10.1038/s41467-023-43875-y
Rho GTPase activity crosstalk mediated by Arhgef11 and Arhgef12 coordinates cell protrusion-retraction cycles
Abstract
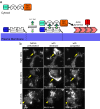
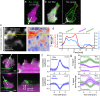
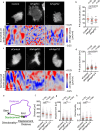
Rho GTPases play a key role in the spatio-temporal coordination of cytoskeletal dynamics during cell migration. Here, we directly investigate crosstalk between the major Rho GTPases Rho, Rac and Cdc42 by combining rapid activity perturbation with activity measurements in mammalian cells. These studies reveal that Rac stimulates Rho activity. Direct measurement of spatio-temporal activity patterns show that Rac activity is tightly and precisely coupled to local cell protrusions, followed by Rho activation during retraction. Furthermore, we find that the Rho-activating Lbc-type GEFs Arhgef11 and Arhgef12 are enriched at transient cell protrusions and retractions and recruited to the plasma membrane by active Rac. In addition, their depletion reduces activity crosstalk, cell protrusion-retraction dynamics and migration distance and increases migration directionality. Thus, our study shows that Arhgef11 and Arhgef12 facilitate exploratory cell migration by coordinating cell protrusion and retraction by coupling the activity of the associated regulators Rac and Rho.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Hodge, R. G. & Ridley, A. J. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol.17, 496–510 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 823/4-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 823/8-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 823/9-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 823/10-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 0315258/Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

