Prion protein conversion at two distinct cellular sites precedes fibrillisation
- PMID: 38102121
- PMCID: PMC10724300
- DOI: 10.1038/s41467-023-43961-1
Prion protein conversion at two distinct cellular sites precedes fibrillisation
Abstract
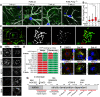
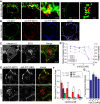
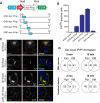
The self-templating nature of prions plays a central role in prion pathogenesis and is associated with infectivity and transmissibility. Since propagation of proteopathic seeds has now been acknowledged a principal pathogenic process in many types of dementia, more insight into the molecular mechanism of prion replication is vital to delineate specific and common disease pathways. By employing highly discriminatory anti-PrP antibodies and conversion-tolerant PrP chimera, we here report that de novo PrP conversion and formation of fibril-like PrP aggregates are distinct in mechanistic and kinetic terms. De novo PrP conversion occurs within minutes after infection at two subcellular locations, while fibril-like PrP aggregates are formed exclusively at the plasma membrane, hours after infection. Phenotypically distinct pools of abnormal PrP at perinuclear sites and the plasma membrane show differences in N-terminal processing, aggregation state and fibril formation and are linked by exocytic transport via synaptic and large-dense core vesicles.
© 2023. Crown.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Prusiner, S. B. Molecular biology and genetics of prion diseases. Cold Spring Harb. Symp. Quant. Biol.61, 473–493 (1996). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

