An immunoinformatics study reveals a new BoLA-DR-restricted CD4+ T cell epitopes on the Gag protein of bovine leukemia virus
- PMID: 38102157
- PMCID: PMC10724172
- DOI: 10.1038/s41598-023-48899-4
An immunoinformatics study reveals a new BoLA-DR-restricted CD4+ T cell epitopes on the Gag protein of bovine leukemia virus
Abstract
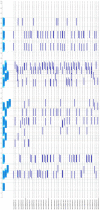
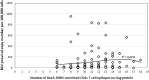
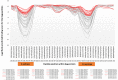
Bovine leukemia virus (BLV) is the causative agent of enzootic bovine leucosis (EBL), which has been reported worldwide. The expression of viral structural proteins: surface glycoprotein (gp51) and three core proteins - p15 (matrix), p24 (capsid), and p12 (nucleocapsid) induce a strong humoral and cellular immune response at first step of infection. CD4+ T-cell activation is generally induced by bovine leukocyte antigen (BoLA) region- positive antigen-presenting cells (APC) after processing of an exogenous viral antigen. Limited data are available on the BLV epitopes from the core proteins recognized by CD4+ T-cells. Thus, immunoinformatic analysis of Gag sequences obtained from 125 BLV isolates from Poland, Canada, Pakistan, Kazakhstan, Moldova and United States was performed to identify the presence of BoLA-DRB3 restricted CD4+ T-cell epitopes. The 379 15-mer overlapping peptides spanning the entire Gag sequence were run in BoLA-DRB3 allele-binding regions using a BoLA-DRB- peptide binding affinity prediction algorithm. The analysis identified 22 CD4+ T-cell peptide epitopes of variable length ranging from 17 to 22 amino acids. The predicted epitopes interacted with 73 different BoLA-DRB3 alleles found in BLV-infected cattle. Importantly, two epitopes were found to be linked with high proviral load in PBMC. A majority of dominant and subdominant epitopes showed high conservation across different viral strains, and therefore could be attractive targets for vaccine development.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
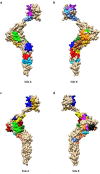


References
-
- Schwartz I, Levy D. Pathobiology of bovine leukemia virus. Vet. Res. 1994;25:521–536. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

