Trypanosome diversity in small mammals in Uganda and the spread of Trypanosoma lewisi to native species
- PMID: 38102492
- PMCID: PMC10724337
- DOI: 10.1007/s00436-023-08048-2
Trypanosome diversity in small mammals in Uganda and the spread of Trypanosoma lewisi to native species
Abstract
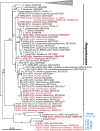
Uganda's diverse small mammalian fauna thrives due to its rich habitat diversity, which hosts a wide range of blood parasites, including trypanosomes, particularly the subgenus Herpetosoma typical for rodent hosts. We screened a total of 711 small mammals from various habitats for trypanosomes, with 253 microscopically examined blood smears and 458 tissue samples tested by nested PCR of the 18S rRNA gene. Of 51 rodent and 12 shrew species tested, microscopic screening reaches 7% overall prevalence (with four rodent species positive out of 15 and none of the shrew species out of four), while nested PCR indicated a prevalence of 13% (17 rodent and five shrew species positive out of 49 and 10, respectively). We identified 27 genotypes representing 11 trypanosome species, of which the majority (24 genotypes/9 species) belong to the Herpetosoma subgenus. Among these, we detected 15 new genotypes and two putative new species, labeled AF24 (found in Lophuromys woosnami) and AF25 (in Graphiurus murinus). Our finding of three new genotypes of the previously detected species AF01 belonging to the subgenus Ornithotrypanum in two Grammomys species and Oenomys hypoxanthus clearly indicates the consistent occurrence of this avian trypanosome in African small mammals. Additionally, in Aethomys hindei, we detected the putative new species of the subgenus Aneza. Within the T. lewisi subclade, we detected eleven genotypes, including six new; however, only the genotype AF05b from Mus and Rattus represents the invasive T. lewisi. Our study has improved our understanding of trypanosome diversity in African small mammals. The detection of T. lewisi in native small mammals expands the range of host species and highlighting the need for a broader approach to the epidemiology of T. lewisi.
Keywords: Aneza; Herpetosoma; Muridae; Ornithotrypanum; Soricidae.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Alsarraf M, Bednarska M, Mohallal EM, Mierzejewska EJ, Behnke-Borowczyk J, Zalat S, Gilbert F, Welc-Falęciak R, Kloch A, Behnke JM, Bajer A (2016) Long-term spatiotemporal stability and dynamic changes in the haemoparasite community of spiny mice (Acomys dimidiatus) in four montane wadis in the St. Katherine Protectorate, Sinai Egypt. Parasit Vectors 9:195. 10.1007/s00248-014-0390-9 - PMC - PubMed
-
- Bryja J, Mikula O, Šumbera R, Meheretu Y, Aghova T, Lavrenchenko L, Mazoch V, Oguge N, Mbau SJ, Welegerima K, Amundala N, Colyn M, Leirs H, Verheyen E. Pan-African phylogeny of Mus (subgenus Nannomys) reveals one of the most successful mammal radiations in Africa. BMC Evol Biol. 2014;14:256. doi: 10.1186/s12862-014-0256-2. - DOI - PMC - PubMed
-
- Chao A, Chiu CH (2016) Species richness: estimation and comparison. Wiley StatsRef: Statistics Reference Online. 10.1002/9781118445112.stat03432.pub2
MeSH terms
Grants and funding
- 20-07091J/The Czech Science Foundation
- 20-07091J/The Czech Science Foundation
- 20-07091J/The Czech Science Foundation
- 23-06116S/Czech Science Foundation
- 23-06116S/Czech Science Foundation
- 23-06116S/Czech Science Foundation
- 5799-TZ/Africa Centre of Excellence for Innovative Rodent Pest Management and Biosensor Technology Development (ACE IRPM & BTD)
- 5799-TZ/Africa Centre of Excellence for Innovative Rodent Pest Management and Biosensor Technology Development (ACE IRPM & BTD)
- 5799-TZ/Africa Centre of Excellence for Innovative Rodent Pest Management and Biosensor Technology Development (ACE IRPM & BTD)
- 5799-TZ/Africa Centre of Excellence for Innovative Rodent Pest Management and Biosensor Technology Development (ACE IRPM & BTD)
- 5799-TZ/Africa Centre of Excellence for Innovative Rodent Pest Management and Biosensor Technology Development (ACE IRPM & BTD)
LinkOut - more resources
Full Text Sources

