Validation of method for faecal sampling in cats and dogs for faecal microbiome analysis
- PMID: 38102642
- PMCID: PMC10724939
- DOI: 10.1186/s12917-023-03842-7
Validation of method for faecal sampling in cats and dogs for faecal microbiome analysis
Abstract
Background: Reproducible and reliable studies of cat and dog faecal microbiomes are dependent on many methodology-based variables including how the faecal stools are sampled and stored prior to processing. The current study aimed to establish an appropriate method for sampling and storing faecal stools from cats and dogs which may also be applied to privately-owned pets. The approach investigated the effects of storing faeces for up to 12 h at room temperature and sampling from various locations within the stool in terms of microbial diversity, relative taxa abundances and DNA yield. Faeces were collected from 10 healthy cats and 10 healthy dogs and stored at room temperature (20 °C). Samples were taken from various locations within the stool (the first emitted part (i), the middle (ii) and the last emitted end (iii), at either surface or core) at 0, 0.5, 1, 2, 3, 6 and 12 h, stabilised and stored at -80 °C. DNA was extracted from all samples, using Illumina NovaSeq.
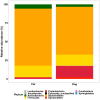
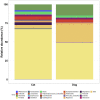
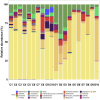
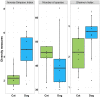
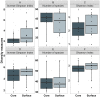
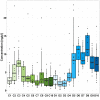
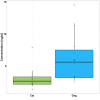
Results: Faecal bacterial composition of dogs and cats shown no statistically significant differences in alpha diversity. Bacteroidetes, Firmicutes, Proteobacteria and Actinobacteria were the most prevalent phyla. Cat and dog samples were characterized by a dominance of Prevotella, and a lack of Fusobacterium in feline stools. Room temperature storage of cat and dog faecal samples generally had no significant effect on alpha diversity, relative taxa abundance or DNA yield for up to 12 h. Sampling from regions i, ii or iii of the stool at the surface or core did not significantly influence the outcome. However, surface cat faecal samples stored at room temperature for 12 h showed a significant increase in two measures of alpha diversity and there was a tendency for a similar effect in dogs. When comparing samples with beta diversity measures, it appeared that for dog and cat samples, individual effect has the strongest impact on the observed microbial diversity (R2 0.64 and 0.88), whereas sampling time, depth and horizontal locations significantly affected the microbial diversity but with less impact.
Conclusion: Cat and dog faeces were stable at room temperature for up to 12 h, with no significant changes in alpha diversity, relative taxa abundance and DNA concentration. Beta diversity analysis demonstrated that despite an impact of the sampling storing time and the surface of the sampling, we preserved the identity of the microbial structure linked to the individual. Finally, the data suggest that faecal stools stored for > 6 h at room temperature should be sampled at the core, not the surface.
Keywords: Cat; Dog; Faeces; Gastrointestinal; Microbiome; Storage; Temperature.
© 2023. The Author(s).
Conflict of interest statement
Xavier Langon is an employee of Royal Canin, with an interest in petfood manufacture.
Figures


Similar articles
-
Storage and handling of human faecal samples affect the gut microbiome composition: A feasibility study.J Microbiol Methods. 2019 Sep;164:105668. doi: 10.1016/j.mimet.2019.105668. Epub 2019 Jul 11. J Microbiol Methods. 2019. PMID: 31302202
-
Effects of a milk oligosaccharide biosimilar on fecal characteristics, microbiota, and bile acid, calprotectin, and immunoglobulin concentrations of healthy adult dogs treated with metronidazole.J Anim Sci. 2023 Jan 3;101:skad011. doi: 10.1093/jas/skad011. J Anim Sci. 2023. PMID: 36617268 Free PMC article.
-
The effects of galacto-oligosaccharides on faecal parameters in healthy dogs and cats.Res Vet Sci. 2024 Feb;167:105116. doi: 10.1016/j.rvsc.2023.105116. Epub 2023 Dec 28. Res Vet Sci. 2024. PMID: 38160491
-
Catching a glimpse of the bacterial gut community of companion animals: a canine and feline perspective.Microb Biotechnol. 2020 Nov;13(6):1708-1732. doi: 10.1111/1751-7915.13656. Epub 2020 Aug 30. Microb Biotechnol. 2020. PMID: 32864871 Free PMC article. Review.
-
The effects of raw-meat diets on the gastrointestinal microbiota of the cat and dog: a review.N Z Vet J. 2022 Jan;70(1):1-9. doi: 10.1080/00480169.2021.1975586. Epub 2021 Sep 19. N Z Vet J. 2022. PMID: 34463606 Review.
Cited by
-
Feline enteropathogens and molecular diagnostics: benefits, limitations and clinical applications.J Feline Med Surg. 2025 Aug;27(8):1098612X251352746. doi: 10.1177/1098612X251352746. Epub 2025 Aug 8. J Feline Med Surg. 2025. PMID: 40776806 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

