IDR-targeting compounds suppress HPV genome replication via disruption of phospho-BRD4 association with DNA damage response factors
- PMID: 38103559
- PMCID: PMC10843765
- DOI: 10.1016/j.molcel.2023.11.022
IDR-targeting compounds suppress HPV genome replication via disruption of phospho-BRD4 association with DNA damage response factors
Abstract
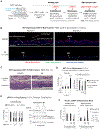
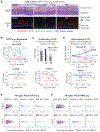
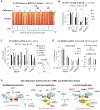
Compounds binding to the bromodomains of bromodomain and extra-terminal (BET) family proteins, particularly BRD4, are promising anticancer agents. Nevertheless, side effects and drug resistance pose significant obstacles in BET-based therapeutics development. Using high-throughput screening of a 200,000-compound library, we identified small molecules targeting a phosphorylated intrinsically disordered region (IDR) of BRD4 that inhibit phospho-BRD4 (pBRD4)-dependent human papillomavirus (HPV) genome replication in HPV-containing keratinocytes. Proteomic profiling identified two DNA damage response factors-53BP1 and BARD1-crucial for differentiation-associated HPV genome amplification. pBRD4-mediated recruitment of 53BP1 and BARD1 to the HPV origin of replication occurs in a spatiotemporal and BRD4 long (BRD4-L) and short (BRD4-S) isoform-specific manner. This recruitment is disrupted by phospho-IDR-targeting compounds with little perturbation of the global transcriptome and BRD4 chromatin landscape. The discovery of these protein-protein interaction inhibitors (PPIi) not only demonstrates the feasibility of developing PPIi against phospho-IDRs but also uncovers antiviral agents targeting an epigenetic regulator essential for virus-host interaction and cancer development.
Keywords: 53BP1; BARD1; BET; BRD4; DDR; HPV; IDR; PPI inhibitors; antiviral; compound.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

