Genome-wide association study meta-analysis of blood pressure traits and hypertension in sub-Saharan African populations: an AWI-Gen study
- PMID: 38104120
- PMCID: PMC10725455
- DOI: 10.1038/s41467-023-44079-0
Genome-wide association study meta-analysis of blood pressure traits and hypertension in sub-Saharan African populations: an AWI-Gen study
Abstract
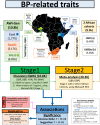
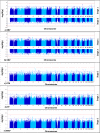
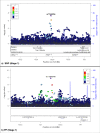
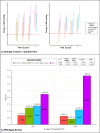
Most hypertension-related genome-wide association studies (GWASs) focus on non-African populations, despite hypertension (a major risk factor for cardiovascular disease) being highly prevalent in Africa. The AWI-Gen study GWAS meta-analysis for blood pressure (BP)-related traits (systolic and diastolic BP, pulse pressure, mean-arterial pressure and hypertension) from three sub-Saharan African geographic regions (N = 10,775), identifies two novel genome-wide significant signals (p < 5E-08): systolic BP near P2RY1 (rs77846204; intergenic variant, p = 4.95E-08) and pulse pressure near LINC01256 (rs80141533; intergenic variant, p = 1.76E-08). No genome-wide signals are detected for the AWI-Gen GWAS meta-analysis with previous African-ancestry GWASs (UK Biobank (African), Uganda Genome Resource). Suggestive signals (p < 5E-06) are observed for all traits, with 29 SNPs associating with more than one trait and several replicating known associations. Polygenic risk scores (PRSs) developed from studies on different ancestries have limited transferability, with multi-ancestry PRS providing better prediction. This study provides insights into the genetics of BP variation in African populations.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Update of
-
Genome-wide Association Study Meta-analysis of Blood Pressure Traits and Hypertension in Sub-Saharan African Populations: An AWI-Gen Study.Res Sq [Preprint]. 2023 Feb 13:rs.3.rs-2532794. doi: 10.21203/rs.3.rs-2532794/v1. Res Sq. 2023. Update in: Nat Commun. 2023 Dec 16;14(1):8376. doi: 10.1038/s41467-023-44079-0. PMID: 36824767 Free PMC article. Updated. Preprint.
References
-
- WHO. World health statistics 2022: monitoring health for the SDGs, sustainable development goals (World Health Organization, 2022).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

