Local energetic frustration conservation in protein families and superfamilies
- PMID: 38104123
- PMCID: PMC10725452
- DOI: 10.1038/s41467-023-43801-2
Local energetic frustration conservation in protein families and superfamilies
Abstract
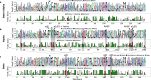
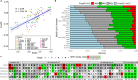
Energetic local frustration offers a biophysical perspective to interpret the effects of sequence variability on protein families. Here we present a methodology to analyze local frustration patterns within protein families and superfamilies that allows us to uncover constraints related to stability and function, and identify differential frustration patterns in families with a common ancestry. We analyze these signals in very well studied protein families such as PDZ, SH3, ɑ and β globins and RAS families. Recent advances in protein structure prediction make it possible to analyze a vast majority of the protein space. An automatic and unsupervised proteome-wide analysis on the SARS-CoV-2 virus demonstrates the potential of our approach to enhance our understanding of the natural phenotypic diversity of protein families beyond single protein instances. We apply our method to modify biophysical properties of natural proteins based on their family properties, as well as perform unsupervised analysis of large datasets to shed light on the physicochemical signatures of poorly characterized proteins such as the ones belonging to emergent pathogens.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

